7LFQ
 
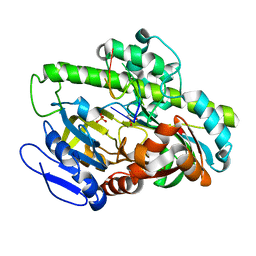 | | Pyrococcus RNA ligase | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*CP*C)-3'), POTASSIUM ION, RNA-splicing ligase RtcB, ... | | Authors: | Goldgur, Y, Shuman, S, Banerjee, A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of 3'-PO 4 /5'-OH RNA ligase RtcB in complex with a 5'-OH oligonucleotide.
Rna, 27, 2021
|
|
4RKG
 
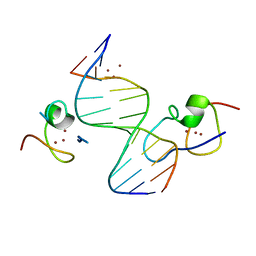 | | Structure of the MSL2 CXC domain bound with a non-specific (GC)6 DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), E3 ubiquitin-protein ligase msl-2, ZINC ION | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
6R8Z
 
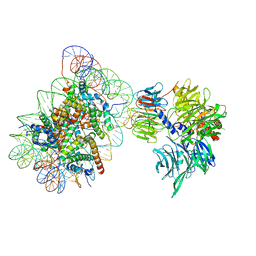 | | Cryo-EM structure of NCP_THF2(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R90
 
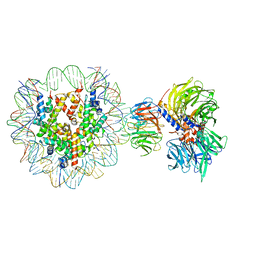 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class A | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R8Y
 
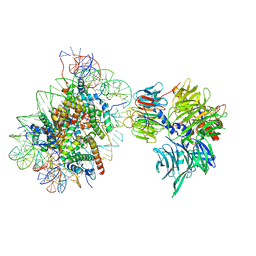 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6R91
 
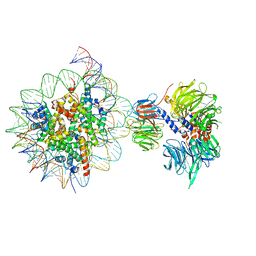 | | Cryo-EM structure of NCP_THF2(-3)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
2ZKD
 
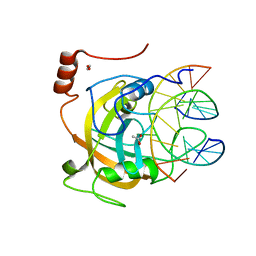 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), ... | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
6M2V
 
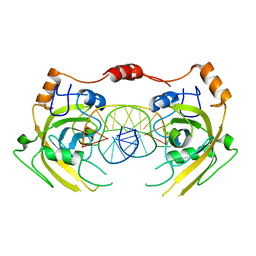 | | Crystal structure of UHRF1 SRA complexed with fully-mCHG DNA. | | Descriptor: | DNA (5'-D(*TP*CP*AP*CP*GP*(5CM)P*TP*GP*CP*GP*TP*GP*A)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Abhishek, S, Nakarakanti, N.K, Deeksha, W, Rajakumara, E. | | Deposit date: | 2020-03-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into recognition of symmetric methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA.
Int.J.Biol.Macromol., 170, 2021
|
|
6R92
 
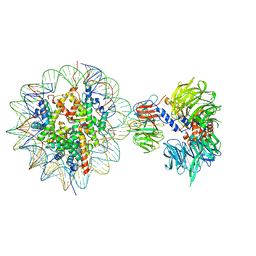 | | Cryo-EM structure of NCP-THF2(+1)-UV-DDB class B | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
5CKK
 
 | |
7K01
 
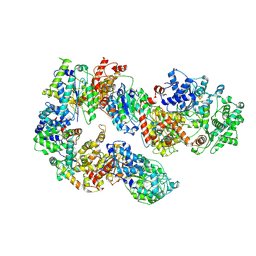 | | Structure of TFIIH in TFIIH/Rad4-Rad23-Rad33 DNA opening complex | | Descriptor: | DNA repair helicase RAD25, DNA repair helicase RAD3, General transcription and DNA repair factor IIH subunit SSL1, ... | | Authors: | van Eeuwen, T, Min, J.H, Murakami, K. | | Deposit date: | 2020-09-02 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
8EWV
 
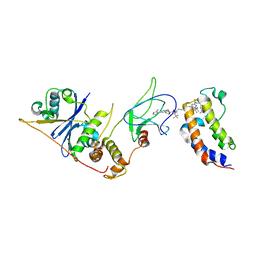 | | DNA-encoded library (DEL)-enabled discovery of proximity inducing small molecules | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Schreiber, S.L, Shu, W, Ma, X, Michaud, G, Bonazzi, S, Berst, F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | DNA-encoded library-enabled discovery of proximity-inducing small molecules.
Nat.Chem.Biol., 20, 2024
|
|
4CI3
 
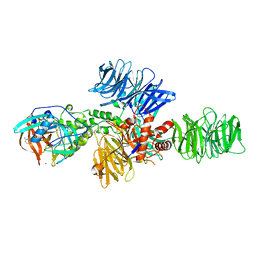 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4CI2
 
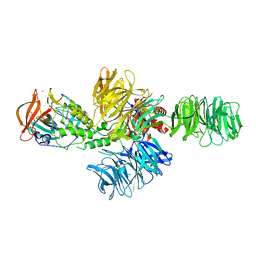 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to lenalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Lenalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
3RCZ
 
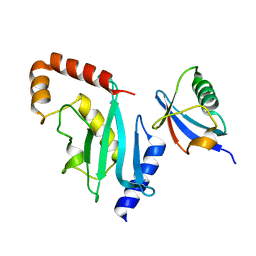 | | Rad60 SLD2 Ubc9 Complex | | Descriptor: | DNA repair protein rad60, SUMO-conjugating enzyme ubc9 | | Authors: | Perry, J.J.P, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA repair and global sumoylation are regulated by distinct Ubc9 noncovalent complexes.
Mol.Cell.Biol., 31, 2011
|
|
6Q00
 
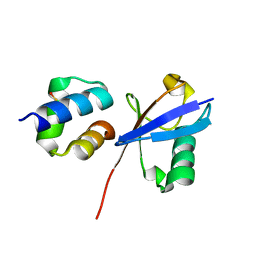 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 1 | | Descriptor: | POTASSIUM ION, Tyrosyl-DNA phosphodiesterase 2, Ubiquitin | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
6WGC
 
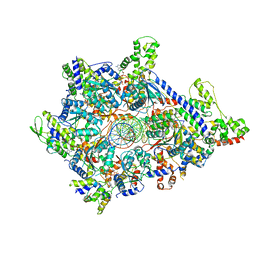 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3SR2
 
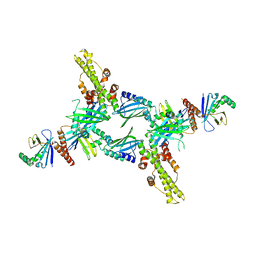 | | Crystal Structure of Human XLF-XRCC4 Complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Hammel, M, Classen, S, Tainer, J.A. | | Deposit date: | 2011-07-06 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.9708 Å) | | Cite: | XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
J.Biol.Chem., 286, 2011
|
|
6WGG
 
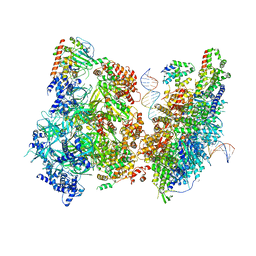 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGI
 
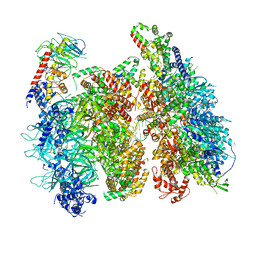 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OEM
 
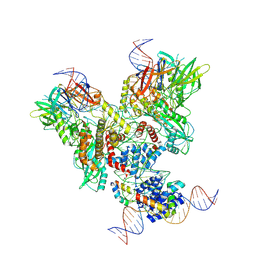 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA0) | | Descriptor: | DNA (46-MER), DNA (57-MER), High mobility group protein B1, ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5NP7
 
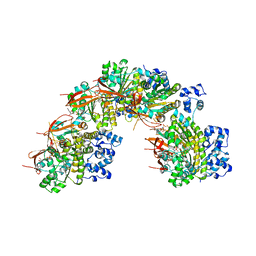 | |
2D8M
 
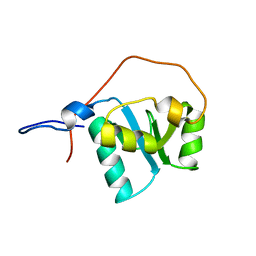 | |
3SZQ
 
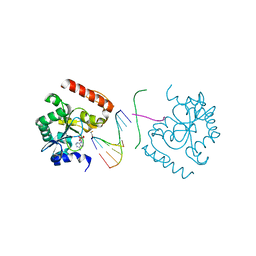 | | Structure of an S. pombe APTX/DNA/AMP/Zn complex | | Descriptor: | 5'-D(*CP*CP*CP*TP*G)-3', 5'-D(*TP*AP*TP*CP*GP*GP*AP*AP*TP*CP*AP*GP*GP*G)-3', ADENOSINE MONOPHOSPHATE, ... | | Authors: | Tumbale, P, Krahn, J, Williams, R.S. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3Q4F
 
 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
