4N3B
 
 | | Crystal Structure of human O-GlcNAc Transferase bound to a peptide from HCF-1 pro-repeat2(1-26)E10Q and UDP-5SGlcNAc | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Host cell factor 1, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Lazarus, M.B, Herr, W, Walker, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | HCF-1 is cleaved in the active site of O-GlcNAc transferase.
Science, 342, 2013
|
|
2CFI
 
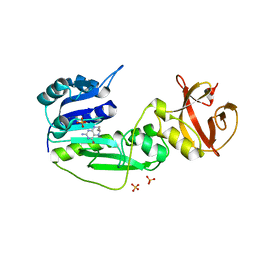 | | The hydrolase domain of human 10-FTHFD in complex with 6- formyltetrahydropterin | | Descriptor: | 10-FORMYLTETRAHYDROFOLATE DEHYDROGENASE, 6-FORMYLTETRAHYDROPTERIN, SULFATE ION | | Authors: | Kursula, P, Stenmark, P, Arrowsmith, C, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Ogg, D.J, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Weigelt, J. | | Deposit date: | 2006-02-21 | | Release date: | 2006-03-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the Hydrolase Domain of Human 10-Formyltetrahydrofolate Dehydrogenase and its Complex with a Substrate Analogue.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4MV8
 
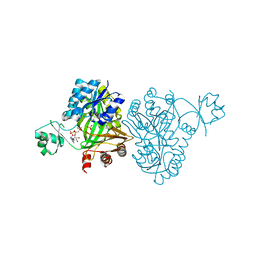 | | Crystal Structure of Biotin Carboxylase from Haemophilus influenzae in Complex with AMPPCP and Phosphate | | Descriptor: | Biotin carboxylase, PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R.J, Waldrop, G.L. | | Deposit date: | 2013-09-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4N49
 
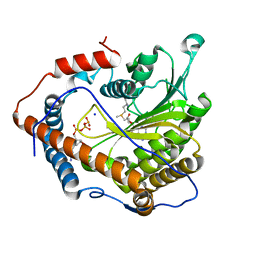 | | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1 Protein in complex with m7GpppG and SAM | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Smietanski, M, Werener, M, Purta, E, Kaminska, K.H, Stepinski, J, Darzynkiewicz, E, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation.
Nat Commun, 5, 2014
|
|
1NHQ
 
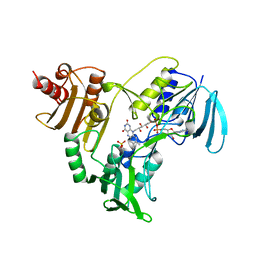 | | CRYSTALLOGRAPHIC ANALYSES OF NADH PEROXIDASE CYS42ALA AND CYS42SER MUTANTS: ACTIVE SITE STRUCTURE, MECHANISTIC IMPLICATIONS, AND AN UNUSUAL ENVIRONMENT OF ARG303 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE, SULFATE ION | | Authors: | Mande, S.S, Claiborne, A, Hol, W.G.J. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analyses of NADH peroxidase Cys42Ala and Cys42Ser mutants: active site structures, mechanistic implications, and an unusual environment of Arg 303.
Biochemistry, 34, 1995
|
|
3CA3
 
 | | Crystal structure of Sambucus Nigra Agglutinin II (SNA-II)-tetragonal crystal form- complexed to N-Acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
4MVX
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-phenylurea (Chem 1356) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-phenylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
7JVL
 
 | |
4MWC
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[(2-methyl-1-benzothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1540) | | Descriptor: | 1-(3-{[(2-methyl-1-benzothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
3CAI
 
 | | Crystal structure of Mycobacterium tuberculosis Rv3778c protein | | Descriptor: | POSSIBLE AMINOTRANSFERASE | | Authors: | Covarrubias, A.S, Larsson, A.M, Jones, T.A, Bergfors, T, Mowbray, S.L, Unge, T. | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biochemical function studies of Mycobacterium tuberculosis essential protein Rv3778c
To be published
|
|
7RAS
 
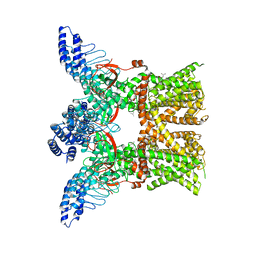 | | Structure of TRPV3 in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-07-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the plant-derived coumarin osthole.
Embo Rep., 22, 2021
|
|
4MWZ
 
 | |
8W1C
 
 | | Cryo-EM structure of BTV pre-subcore | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
7RAU
 
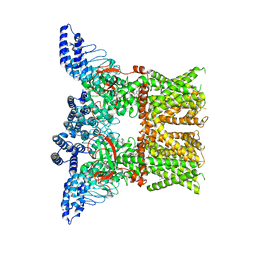 | | Structure of TRPV3 in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-07-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the plant-derived coumarin osthole.
Embo Rep., 22, 2021
|
|
3FYG
 
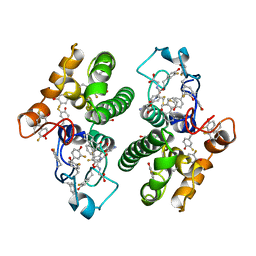 | | CRYSTAL STRUCTURE OF TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, MU CLASS TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE OF ISOENZYME | | Authors: | Xiao, G, Parsons, J.F, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-08-07 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational changes in the crystal structure of rat glutathione transferase M1-1 with global substitution of 3-fluorotyrosine for tyrosine.
J.Mol.Biol., 281, 1998
|
|
7RL1
 
 | | AAVrh.10-7x capsid | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural Study of Aavrh.10 Receptor and Antibody Interactions.
J.Virol., 95, 2021
|
|
2CJL
 
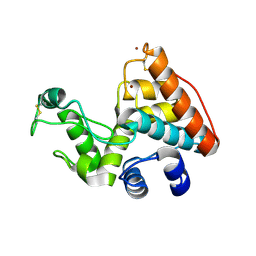 | | CRYSTAL STRUCTURE AND ENZYMATIC PROPERTIES OF A BACTERIAL FAMILY 19 CHITINASE REVEAL DIFFERENCES WITH PLANT ENZYMES | | Descriptor: | SECRETED CHITINASE, ZINC ION | | Authors: | Hoell, I.A, Dalhus, B, Heggset, E.B, Aspmo, S.I, Eijsink, V.G.H. | | Deposit date: | 2006-04-04 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Enzymatic Properties of a Bacterial Family 19 Chitinase Reveal Differences with Plant Enzymes
FEBS J., 273, 2006
|
|
8WA2
 
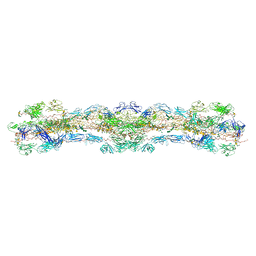 | | cryo-EM structure of native mastigonemes isolated from Chlamydomonas reinhardtii at 3.0 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, J, Tao, H, Chen, J, Pan, J, Yan, C, Yan, N. | | Deposit date: | 2023-09-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-guided discovery of protein and glycan components in native mastigonemes.
Cell, 187, 2024
|
|
4H19
 
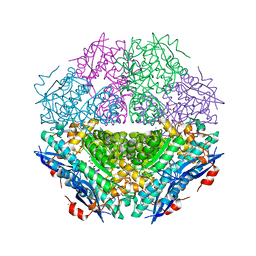 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502087) from agrobacterium tumefaciens, with bound Mg and d-ribonohydroxamate, ordered loop | | Descriptor: | (2R,3R,4R)-N,2,3,4,5-pentakis(oxidanyl)pentanamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502087) from agrobacterium tumefaciens, with bound Mg and d-ribonohydroxamate, ordered loop
To be Published
|
|
2CK1
 
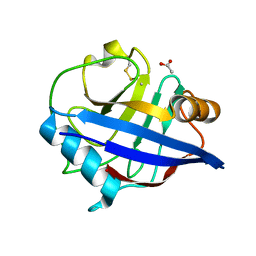 | | The structure of oxidised cyclophilin A from s. mansoni | | Descriptor: | ACETATE ION, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE E | | Authors: | Gourlay, L.J, Angelucci, F, Bellelli, A, Boumis, G, Miele, A.E, Brunori, M. | | Deposit date: | 2006-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-Dimensional Structure of Two Redox States of Cyclophilin-A from Schistosoma Mansoni: Evidence for Redox- Regulation of Peptidyl-Prolyl Cis-Trans Isomerase Activity.
J.Biol.Chem., 282, 2007
|
|
4MYV
 
 | | Free HSV-2 gD structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
2V3Q
 
 | | Serendipitous discovery and X-ray structure of a human phosphate binding apolipoprotein | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, HUMAN PHOSPHATE BINDING PROTEIN, ... | | Authors: | Morales, R, Berna, A, Carpentier, P, Elias, M, Contreras-Martel, C, Renault, F, Nicodeme, M, Chesne-Seck, M.-L, Bernier, F, Dupuy, J, Schaeffer, C, Diemer, H, Van Dorsselaer, A, Fontecilla, J.C, Masson, P, Rochu, D, Chabriere, E. | | Deposit date: | 2007-06-20 | | Release date: | 2008-07-22 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Tandem Use of X-Ray Crystallography and Mass Spectrometry to Obtain Ab Initio the Complete and Exact Amino Acids Sequence of Hpbp, a Human 38kDa Apolipoprotein
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
8U8O
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 1, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
7RJA
 
 | |
5D7B
 
 | | Trigonal Crystal Structure of an acetylester hydrolase from Corynebacterium glutamicum | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Niefind, K, Toelzer, C, Pal, S, Altenbuchner, J, Watzlawick, H. | | Deposit date: | 2015-08-13 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
