3GKV
 
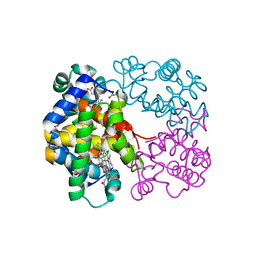 | | X-ray structure of an intermediate along the oxidation pathway of Trematomus bernacchii hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Vergara, A, Mazzarella, L. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Combined crystallographic and spectroscopic analysis of Trematomus bernacchii hemoglobin highlights analogies and differences in the peculiar oxidation pathway of Antarctic fish hemoglobins
Biopolymers, 91, 2009
|
|
8UTK
 
 | | IL-23R minibinder - 23R-B04dslf02IB | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 23R-B04dslf02IB, ... | | Authors: | Bera, A.K, Berger, S.A, Kang, A, Baker, D. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Preclinical proof of principle for orally delivered Th17 antagonist miniproteins.
Cell, 187, 2024
|
|
5KEI
 
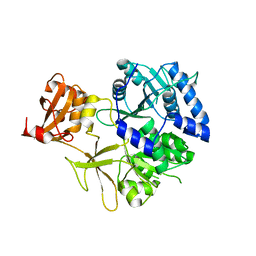 | |
6MYX
 
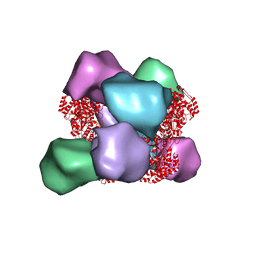 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | Authors: | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
2R4P
 
 | | Crystal structure of the long-chain fatty acid transporter FadL mutant G212E | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2007-08-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Transmembrane passage of hydrophobic compounds through a protein channel wall
Nature, 458, 2009
|
|
6MW3
 
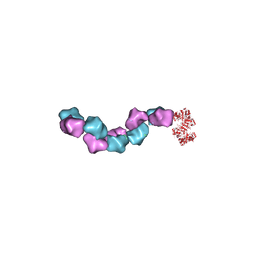 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
4MJH
 
 | |
2R4O
 
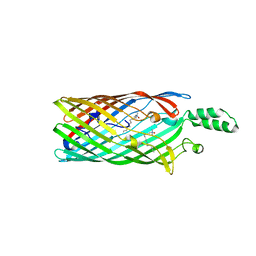 | | Crystal structure of the long-chain fatty acid transporter FadL mutant delta NPA | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, Long-chain fatty acid transport protein | | Authors: | Hearn, E.M, Patel, D.R, Lepore, B.W, Indic, M, van den Berg, B. | | Deposit date: | 2007-08-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transmembrane passage of hydrophobic compounds through a protein channel wall
Nature, 458, 2009
|
|
1DUP
 
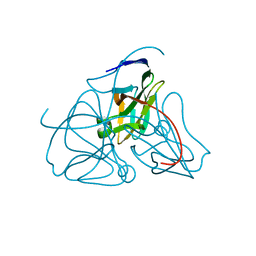 | | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE (D-UTPASE) | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Wilson, K.S, Larsson, G, Nyman, P.O, Cedergren, E. | | Deposit date: | 1995-09-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dUTPase.
Nature, 355, 1992
|
|
1OB1
 
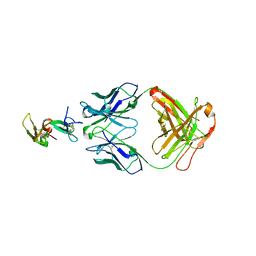 | | Crystal structure of a Fab complex whith Plasmodium falciparum MSP1-19 | | Descriptor: | ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pizarro, J.C, Chitarra, V, Verger, D, Holm, I, Petres, S, Dartville, S, Nato, F, Longacre, S, Bentley, G.A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Fab Complex Formed with Pfmsp1-19, the C-Terminal Fragment of Merozoite Surface Protein 1 from Plasmodium Falciparum: A Malaria Vaccine Candidate
J.Mol.Biol., 328, 2003
|
|
8P8A
 
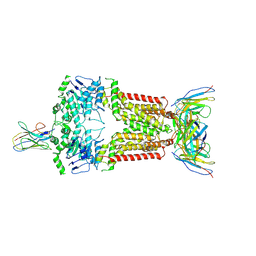 | | Structure of 5D3-Fab and nanobody(Nb17)-bound ABCG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5D3(Fab) heavy chain variable domain, 5D3(Fab) light chain variable domain, ... | | Authors: | Irobalieva, R.N, Manolaridis, I, Jackson, S.M, Ni, D, Pardon, E, Stahlberg, H, Steyaert, J, Locher, K.P. | | Deposit date: | 2023-05-31 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of the Allosteric Inhibition of Human ABCG2 by Nanobodies.
J.Mol.Biol., 435, 2023
|
|
8PAY
 
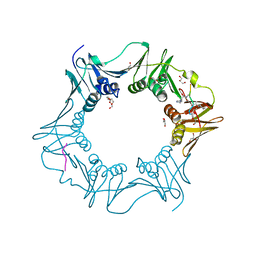 | | Structure of the E.coli DNA polymerase sliding clamp with a covalently bound peptide 2. | | Descriptor: | ACE-GLN-ALC-GLC-LEU-PHE, Beta sliding clamp, GLYCEROL, ... | | Authors: | Compain, G, Monsarrat, C, Blagojevic, J, Brillet, K, Dumas, P, Hammann, P, Kuhn, L, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Burnouf, D, wagner, J, Guichard, G. | | Deposit date: | 2023-06-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Peptide-Based Covalent Inhibitors Bearing Mild Electrophiles to Target a Conserved His Residue of the Bacterial Sliding Clamp.
Jacs Au, 4, 2024
|
|
8ADN
 
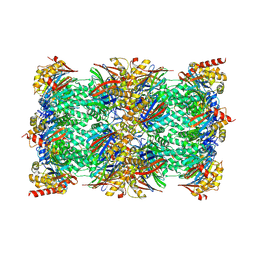 | | Vairimorpha necatrix 20S proteasome from spores | | Descriptor: | Proteasome Inhibitor 31-Like, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Jespersen, N, Ehrenbolger, K, Winiger, R, Svedberg, D, Vossbrinck, C.R, Barandun, J. | | Deposit date: | 2022-07-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the reduced microsporidian proteasome bound by PI31-like peptides in dormant spores.
Nat Commun, 13, 2022
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
8AS5
 
 | |
5O8A
 
 | | Crystal Structure of rsEGFP2 in the non-fluorescent off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
8PM6
 
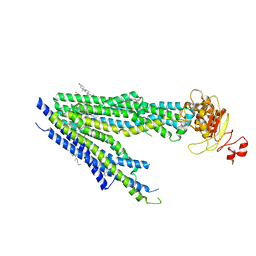 | | Human bile salt export pump (BSEP) in complex with inhibitor GBM in nanodiscs | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Bile salt export pump, CHOLESTEROL | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
5O89
 
 | | Crystal Structure of rsEGFP2 in the fluorescent on-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5O8C
 
 | | Composite structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | Descriptor: | Green fluorescent protein | | Authors: | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2017-06-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
2NPQ
 
 | |
1OQF
 
 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
8PMJ
 
 | | Vanadate-trapped BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, ... | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8W0X
 
 | |
8W0Y
 
 | |
