2MYO
 
 | | SOLUTION STRUCTURE OF MYOTROPHIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MYOTROPHIN | | Authors: | Yang, Y, Nanduri, S, Sen, S, Qin, J. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin.
Structure, 6, 1998
|
|
2N16
 
 | |
2ND7
 
 | | Structure of DK17 in POPC:POPG:Cholesterol:GM1 LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
6UL5
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile (24b), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]-2-fluorobenzonitrile, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Pilch, A, Arnold, E. | | Deposit date: | 2019-10-06 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and Characterization of Fluorine-Substituted Diarylpyrimidine Derivatives as Novel HIV-1 NNRTIs with Highly Improved Resistance Profiles and Low Activity for the hERG Ion Channel.
J.Med.Chem., 63, 2020
|
|
2ND6
 
 | | Structure of DK17 in GM1 LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
7XQ8
 
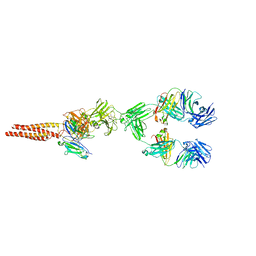 | | Structure of human B-cell antigen receptor of the IgM isotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, ... | | Authors: | Chen, M.Y, Su, Q, Shi, Y.G. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human IgM B cell receptor.
Science, 377, 2022
|
|
2N45
 
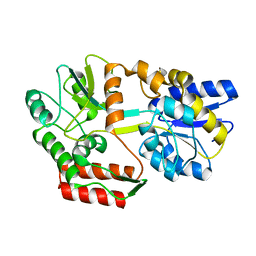 | | EC-NMR Structure of Escherichia coli Maltose-binding protein Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data with a second set of RDC data simulated for an alternative alignment tensor. Northeast Structural Genomics Consortium target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
8KHP
 
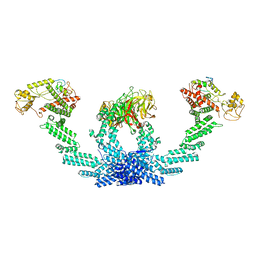 | | CULLIN3-KLHL22-RBX1 E3 ligase | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch-like protein 22 | | Authors: | Su, M.-Y, Su, M.-Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
1G8Y
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC REPLICATIVE HELICASE REPA OF PLASMID RSF1010 | | Descriptor: | REGULATORY PROTEIN REPA | | Authors: | Niedenzu, T, Roeleke, D, Bains, G, Scherzinger, E, Saenger, W. | | Deposit date: | 2000-11-21 | | Release date: | 2001-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010.
J.Mol.Biol., 306, 2001
|
|
2N1B
 
 | |
7Y5Q
 
 | | Structure of 1:1 PAPP-A.STC2 complex(half map) | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
5J36
 
 | | Crystal structure of 60-mer BFDV Capsid Protein | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
1TZV
 
 | | T. maritima NusB, P3121, Form 1 | | Descriptor: | N utilization substance protein B homolog | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
2NSM
 
 | | Crystal structure of the human carboxypeptidase N (Kininase I) catalytic domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase N catalytic chain, SULFATE ION | | Authors: | Keil, C, Maskos, K, Than, M, Hoopes, J.T, Huber, R, Tan, F, Deddish, P.A, Erdoes, E.G, Skidgel, R.A, Bode, W. | | Deposit date: | 2006-11-05 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain
J.Mol.Biol., 366, 2007
|
|
6OFZ
 
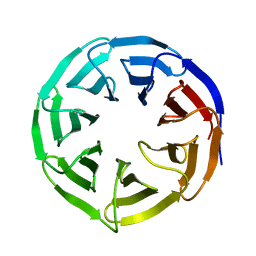 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
2NVW
 
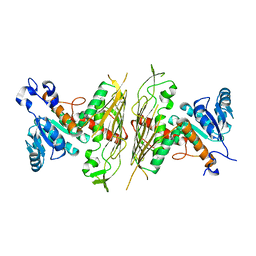 | |
2DA5
 
 | | Solution structure of the second homeobox domain of Zinc fingers and homeoboxes protein 3 (Triple homeobox 1 protein) | | Descriptor: | Zinc fingers and homeoboxes protein 3 | | Authors: | Ohnishi, S, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second homeobox domain of Zinc fingers and homeoboxes protein 3 (Triple homeobox 1 protein)
To be Published
|
|
7XW3
 
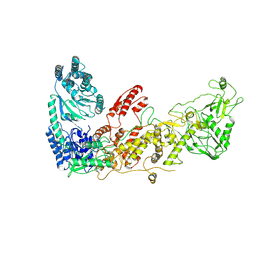 | |
7XW2
 
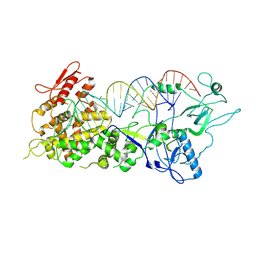 | |
2O8V
 
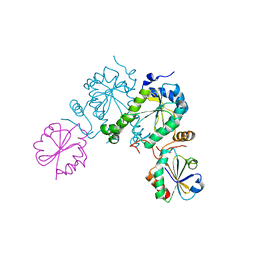 | | PAPS reductase in a covalent complex with thioredoxin C35A | | Descriptor: | Phosphoadenosine phosphosulfate reductase, Thioredoxin 1 | | Authors: | Chartron, J, Shiau, C, Stout, C.D, Carroll, K.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3'-Phosphoadenosine-5'-phosphosulfate Reductase in Complex with Thioredoxin: A Structural Snapshot in the Catalytic Cycle.
Biochemistry, 46, 2007
|
|
2OD9
 
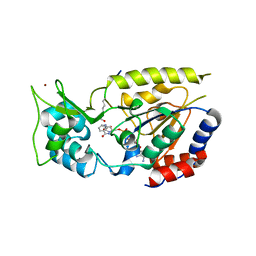 | | Structural Basis for Nicotinamide Inhibition and Base Exchange in Sir2 Enzymes | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, H4 peptide, NAD-dependent deacetylase HST2, ... | | Authors: | Marmorstein, R, Sanders, B.D. | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for nicotinamide inhibition and base exchange in sir2 enzymes.
Mol.Cell, 25, 2007
|
|
7Y09
 
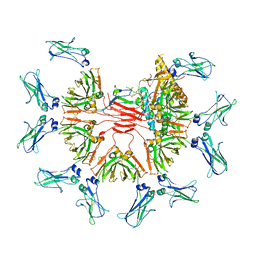 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
7YG2
 
 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DBLMSP2, ... | | Authors: | Shen, H, Ji, C, Xiao, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M.
Nat Commun, 14, 2023
|
|
2NSU
 
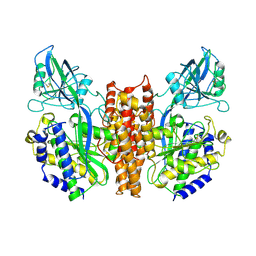 | |
5K14
 
 | | HIV-1 Reverse Transcriptase in complex with a 2,6-difluorophenyl DAPY analog | | Descriptor: | 4-{[4-(2,6-difluoro-4-methoxybenzene-1-carbonyl)pyrimidin-2-yl]amino}benzonitrile, HIV-1 reverse transcriptase (isolate LW123), HIV-1 reverse transcriptase(isolate HXB2) | | Authors: | Lansdon, E.B. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Novel (2,6-difluorophenyl)(2-(phenylamino)pyrimidin-4-yl)methanones with restricted conformation as potent non-nucleoside reverse transcriptase inhibitors against HIV-1.
Eur.J.Med.Chem., 122, 2016
|
|
