6T7I
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CGA inhibitory codon combination. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
8GO8
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GP3
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
6MRC
 
 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
4JQI
 
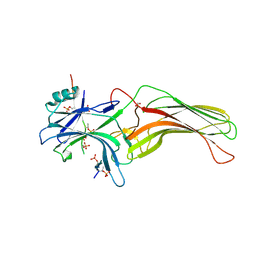 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
8IO5
 
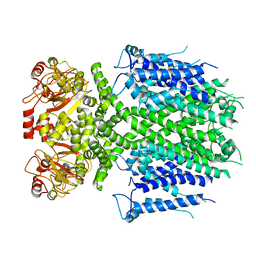 | | Herg1a-herg1b closed state 2 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
8IOB
 
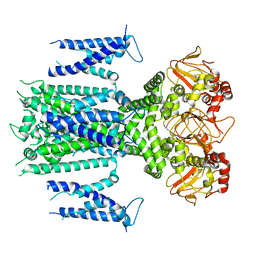 | | Herg1a-herg1b closed state 1 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Herg1a-herg1b closed state 1
To Be Published
|
|
8IO4
 
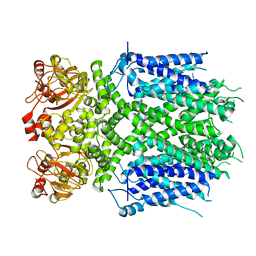 | | Herg1a-herg1b open state | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
8I0N
 
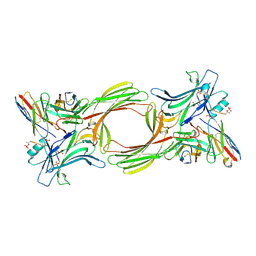 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 (Local refine) | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8I0Q
 
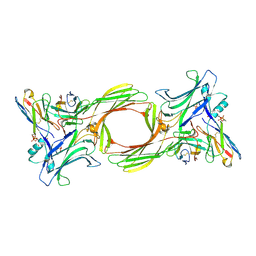 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 (Local refine) | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
4FJ3
 
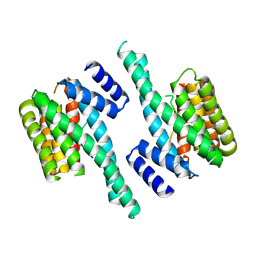 | |
6T83
 
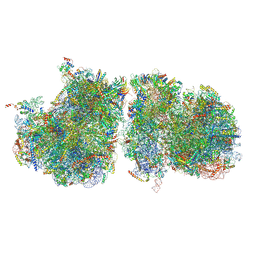 | | Structure of yeast disome (di-ribosome) stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-24 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
4PTE
 
 | | Structure of a carvoxamide compound (15) (N-[4-(ISOQUINOLIN-7-YL)PYRIDIN-2-YL]CYCLOPROPANECARBOXAMIDE) to GSK3b | | Descriptor: | Glycogen synthase kinase-3 beta, N-[4-(isoquinolin-7-yl)pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4N7G
 
 | | Crystal structure of 14-3-3zeta in complex with a peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S, HEXAETHYLENE GLYCOL | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-15 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4PTG
 
 | | Structure of a carboxamine compound (26) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-METHOXYPYRIMIDINE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-{2-[(cyclopropylcarbonyl)amino]pyridin-4-yl}-4-methoxypyrimidine-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4HKC
 
 | |
4PTC
 
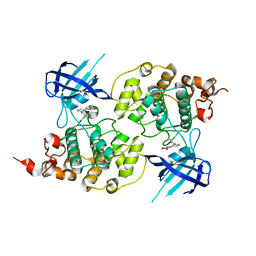 | | Structure of a carboxamide compound (3) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-OXO-4H-1LAMBDA~4~,3-THIAZOLE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-[2-(cyclopropylcarbonylamino)pyridin-4-yl]-4-methoxy-1,3-thiazole-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1DDB
 
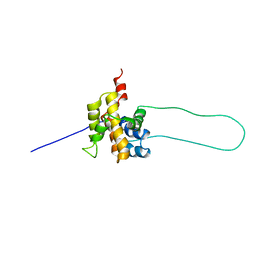 | | STRUCTURE OF MOUSE BID, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (BID) | | Authors: | Mcdonnell, J.M, Fushman, D, Milliman, C, Korsmeyer, S.J, Cowburn, D. | | Deposit date: | 1999-02-19 | | Release date: | 1999-08-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists.
Cell(Cambridge,Mass.), 96, 1999
|
|
5L4K
 
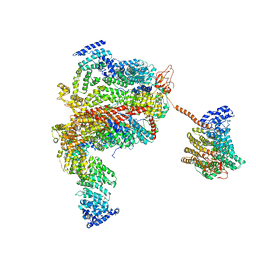 | | The human 26S proteasome lid | | Descriptor: | 26S proteasome complex subunit DSS1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Schweitzer, A, Aufderheide, A, Rudack, T, Beck, F. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the human 26S proteasome at a resolution of 3.9 angstrom.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3J6X
 
 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 5 degree rotation (Class II) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8CDL
 
 | | 80S S. cerevisiae ribosome with ligands in hybrid-2 pre-translocation (PRE-H2) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CDR
 
 | | Translocation intermediate 2 (TI-2) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CCS
 
 | | 80S S. cerevisiae ribosome with ligands in hybrid-1 pre-translocation (PRE-H1) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-27 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
3J78
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class I - non-rotated ribosome with 2 tRNAs) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3J77
 
 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
