8DOK
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
6LUO
 
 | | Structure of nurse shark beta-2-microglobulin | | Descriptor: | Beta-2-microglobulin | | Authors: | Xia, C, Wu, Y. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6ZVU
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174L LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6V1R
 
 | | Crystal structure of iAChSnFR Fluorescent Acetylcholine Sensor precursor binding protein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETYLCHOLINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fan, C, Borden, P.M, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A genetically encoded fluorescent sensor for in vivo acetylcholine detection
To Be Published
|
|
6LUP
 
 | | Crystal structure of shark MHC CLASS I for 2.3 angstrom | | Descriptor: | Beta-2-microglobulin, MHC class I protein, PHE-ALA-ASN-PHE-PHE-ILE-ARG-GLY-LEU | | Authors: | Wu, Y, Xia, C. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6PJN
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3S,5S)-3-hydroxy-5-({1-[(methoxycarbonyl)amino]cyclopentane-1-carbonyl}amino)-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
8E9A
 
 | | Crystal structure of AsfvPolX in complex with 10-23 DNAzyme and Mg | | Descriptor: | DNA/RNA (52-MER), MAGNESIUM ION, Repair DNA polymerase X, ... | | Authors: | Cramer, E.R, Robart, A.R, Kaya, A.I. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of a 10-23 deoxyribozyme exhibiting a homodimer conformation.
Commun Chem, 6, 2023
|
|
6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6V1Z
 
 | | genome-containing AAVrh.39 particles | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
8DP0
 
 | | Structure of p110 gamma bound to the Ras inhibitory nanobody NB7 | | Descriptor: | Nanobody NB7, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Nam, S.E, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Allosteric activation or inhibition of PI3K gamma mediated through conformational changes in the p110 gamma helical domain.
Elife, 12, 2023
|
|
6P07
 
 | | Spastin hexamer in complex with substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sandate, C.R, Szyk, A, Zehr, E, Roll-Mecak, A, Lander, G.C. | | Deposit date: | 2019-05-16 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An allosteric network in spastin couples multiple activities required for microtubule severing.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6ZVD
 
 | |
7M56
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-((3-(3-aminophenethyl)phenoxy)methyl)quinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-({3-[2-(6-aminopyridin-2-yl)ethyl]phenoxy}methyl)quinolin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2021-03-22 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Inhibition of bacterial nitric oxide synthase as an antimicrobial tool in fighting some antibiotic-resistant pathogens
To be published
|
|
6UWK
 
 | |
6M1C
 
 | |
6UWP
 
 | | BACE-1 in complex with compound #32 | | Descriptor: | (1R,2R)-2-[(4aR,7aR)-2-amino-6-(pyrimidin-2-yl)-4a,5,6,7-tetrahydropyrrolo[3,4-d][1,3]thiazin-7a(4H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6ZVV
 
 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-P174W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE
To Be Published
|
|
6LWA
 
 | | Crystal structure of human NEIL1(P2G, E3Q, K242) bound to duplex DNA containing 5-hydroxyuracil (5-OHU) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
8DO4
 
 | |
6P14
 
 | | Structure of spastin AAA domain (T692A mutant) in complex with a diaminotriazole-based inhibitor (crystal form B) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{[5-amino-1-(2-fluoro-6-methoxybenzene-1-carbonyl)-1H-1,2,4-triazol-3-yl]amino}-N-methylbenzamide, ACETATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93001127 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
7ABS
 
 | | Structure of human DCLRE1C/Artemis in complex with DNA - re-evaluation of 6WO0 | | Descriptor: | DNA (5'-D(*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*GP*CP*T)-3'), Protein artemis, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6UX9
 
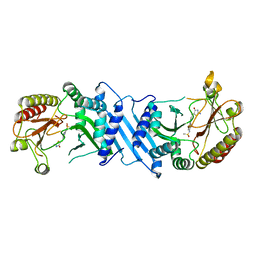 | | Crystal Structure Analysis of PIP4K2A | | Descriptor: | N-[4-(5-{(Z)-[(2E)-2-imino-4-oxo-1,3-thiazolidin-5-ylidene]methyl}pyridin-3-yl)phenyl]methanesulfonamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery and Structure-Activity Relationship Study of ( Z )-5-Methylenethiazolidin-4-one Derivatives as Potent and Selective Pan-phosphatidylinositol 5-Phosphate 4-Kinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6LN0
 
 | |
7M9L
 
 | | HIV-1 Protease WT (NL4-3) in Complex with LR4-15 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-[(2-ethylbutyl){4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}amino]-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | HIV-1 Protease WT (NL4-3) in Complex with LR4-15
To Be Published
|
|
8DPL
 
 | |
