2O3V
 
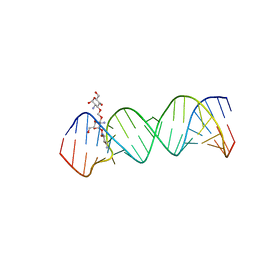 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33 | | Descriptor: | (2S,3R,4R,5S,6R)-3-AMINO-4-({[(2S,3R,4R,5S,6R)-3-AMINO-2-{[(1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYL]OXY}-5-HYDROXY-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-4-YL]OXY}METHOXY)-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-2,5-DIOL, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
2O4K
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, CHLORIDE ION, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
5EF5
 
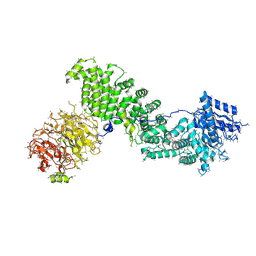 | | Crystal structure of Chaetomium thermophilum Raptor | | Descriptor: | Raptor from Chaetomium thermophilum | | Authors: | Imseng, S, Sauer, E, Aylett, C.H.S, Boehringer, D, Hall, M.N, Ban, N, Maier, T. | | Deposit date: | 2015-10-23 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Architecture of human mTOR complex 1.
Science, 351, 2016
|
|
1E49
 
 | |
1KKE
 
 | |
5A3A
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
2BVG
 
 | |
4ASR
 
 | | Crystal structure of ANCE in complex with Thr6-Bradykinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN-CONVERTING ENZYME, BRADYKININ, ... | | Authors: | Akif, M, Masuyer, G, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2012-05-02 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Peptide Recognition by the Angiotensin-I Converting Enzyme Homologue Ance from Drosophila Melanogaster
FEBS J., 279, 2012
|
|
3N5N
 
 | |
5EHJ
 
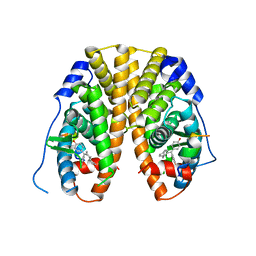 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-[(4aR,8aR)-octahydronaphthalen-2(1H)-ylidenemethanediyl]diphenol | | Descriptor: | 4,4'-[(4aR,8aR)-octahydronaphthalen-2(1H)-ylidenemethanediyl]diphenol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
2OJ0
 
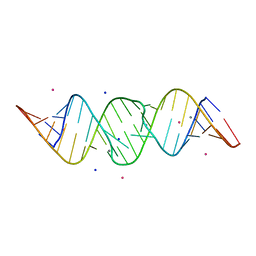 | | Crystal structure of the duplex form of the HIV-1(LAI) RNA dimerization initiation site MN soaked | | Descriptor: | 5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*AP*GP*CP*GP*CP*GP*CP*AP*CP*GP*GP*CP*AP*AP*G)-3', MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2007-01-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cation-dependent cleavage of the duplex form of the subtype-B HIV-1 RNA dimerization initiation site.
Nucleic Acids Res., 38, 2010
|
|
3NKB
 
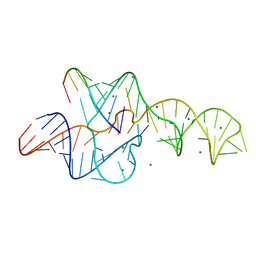 | | A 1.9A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage | | Descriptor: | DNA/RNA (5'-D(*(DUR))-D(*GP*G)-R(P*CP*UP*UP*GP*CP*A)-3'), MAGNESIUM ION, The hepatitis delta virus ribozyme | | Authors: | Chen, J.-H, Yajima, R, Chadalavada, D.M, Chase, E, Bevilacqua, P.C, Golden, B.L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage.
Biochemistry, 49, 2010
|
|
1KOE
 
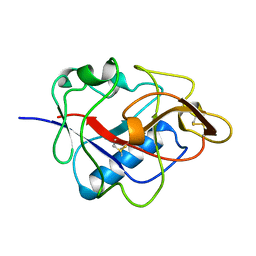 | | ENDOSTATIN | | Descriptor: | ENDOSTATIN | | Authors: | Hohenester, E, Sasaki, T, Olsen, B.R, Timpl, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the angiogenesis inhibitor endostatin at 1.5 A resolution.
EMBO J., 17, 1998
|
|
2VEW
 
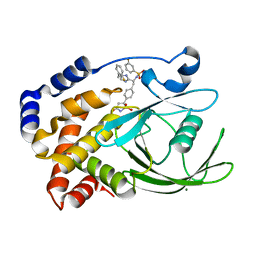 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN ISOTHIAZOLIDINONE-CONTAINING INHIBITOR | | Descriptor: | 3-fluoro-N-[(1S)-1-[4-[(2-fluorophenyl)methyl]imidazol-2-yl]-2-[4-[(5S)-1,1,3-trioxo-1,2-thiazolidin-5-yl]phenyl]ethyl]benzenesulfonamide, MAGNESIUM ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Douty, B, Wayland, B, Ala, P.J, Bower, M.J, Pruitt, J, Bostrom, L, Wei, M, Klabe, R, Gonneville, L, Wynn, R, Burn, T.C, Liu, P.C.C, Combs, A.P, Yue, E.W. | | Deposit date: | 2007-10-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isothiazolidinone Inhibitors of Ptp1B Containing Imidazoles and Imidazolines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2NRT
 
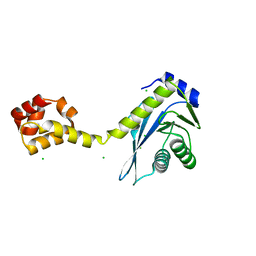 | |
5AEG
 
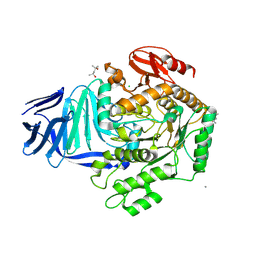 | | A bacterial protein structure in glycoside hydrolase family 31. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-fluoro-alpha-L-idopyranose, ALPHA-GLUCOSIDASE YIHQ, ... | | Authors: | Jin, Y, Speciale, G, Davies, G.J, Williams, S.J, Goddard-Borger, E.D. | | Deposit date: | 2015-08-30 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Yihq is a Sulfoquinovosidase that Cleaves Sulfoquinovosyl Diacylglyceride Sulfolipids.
Nat.Chem.Biol., 12, 2016
|
|
5AFB
 
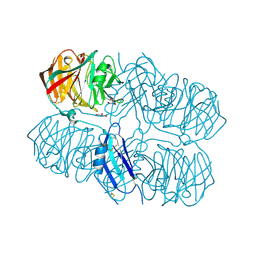 | | Crystal structure of the Latrophilin3 Lectin and Olfactomedin Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Jackson, V.A, del Toro, D, Carrasquero, M, Roversi, P, Harlos, K, Klein, R, Seiradake, E. | | Deposit date: | 2015-01-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis of Latrophilin-Flrt Interaction.
Structure, 23, 2015
|
|
3CWJ
 
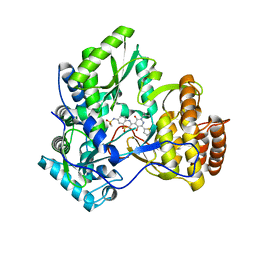 | | Crystal structure of hcv ns5b polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-04-21 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-(1,1-Dioxo-1,4-dihydro-1lambda6-benzo[1,4]thiazin-3-yl)-5-hydroxy-2H-pyridazin-3-ones as potent inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VIX
 
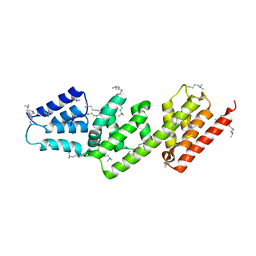 | | Methylated Shigella flexneri MxiC | | Descriptor: | ACETATE ION, GLYCEROL, PROTEIN MXIC | | Authors: | Deane, J.E, Roversi, P, King, C, Johnson, S, Lea, S.M. | | Deposit date: | 2007-12-05 | | Release date: | 2008-03-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of the Shigella Flexneri Type 3 Secretion System Protein Mxic Reveal Conformational Variability Amongst Homologues.
J.Mol.Biol., 377, 2008
|
|
1DT6
 
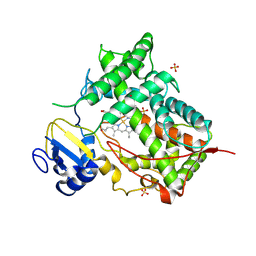 | | STRUCTURE OF MAMMALIAN CYTOCHROME P450 2C5 | | Descriptor: | CYTOCHROME P450 2C5, PROTOPORPHYRIN IX CONTAINING FE, SAMARIUM (III) ION, ... | | Authors: | Williams, P.A, Cosme, J, Sridhar, V, Johnson, E.F, McRee, D.E. | | Deposit date: | 2000-01-11 | | Release date: | 2000-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity.
Mol.Cell, 5, 2000
|
|
5AIQ
 
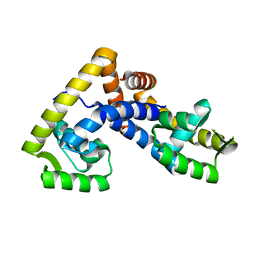 | | Crystal structure of ligand-free NadR | | Descriptor: | TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | Authors: | Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
2VLI
 
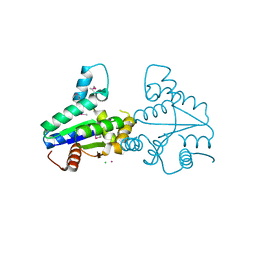 | | Structure of Deinococcus radiodurans tunicamycin resistance protein | | Descriptor: | ANTIBIOTIC RESISTANCE PROTEIN, CADMIUM ION, CHLORIDE ION | | Authors: | Macedo, S, Kapp, U, Leiros, I, Hall, D.R, Mitchell, E. | | Deposit date: | 2008-01-15 | | Release date: | 2008-06-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Deinococcus Radiodurans Tunicamycin-Resistance Protein (Tmrd), a Phosphotransferase.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1KOS
 
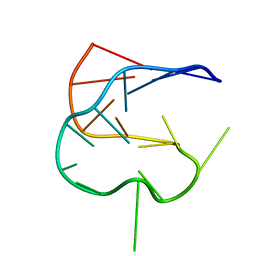 | | SOLUTION NMR STRUCTURE OF AN ANALOG OF THE YEAST TRNA PHE T STEM LOOP CONTAINING RIBOTHYMIDINE AT ITS NATURALLY OCCURRING POSITION | | Descriptor: | 5'-R(*CP*UP*GP*UP*GP*(5MU)P*UP*CP*GP*AP*UP*(CH)P*CP*AP*CP*AP*G)- 3' | | Authors: | Koshlap, K.M, Guenther, R, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A distinctive RNA fold: the solution structure of an analogue of the yeast tRNAPhe T Psi C domain.
Biochemistry, 38, 1999
|
|
4B0T
 
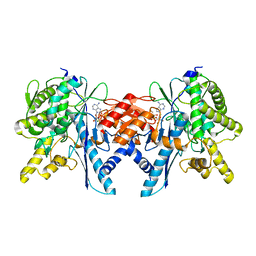 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
2BL2
 
 | | The membrane rotor of the V-type ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, SODIUM ION, UNDECYL-MALTOSIDE, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2005-02-25 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Rotor of the Vacuolar-Type Na- ATPase from Enterococcus Hirae
Science, 308, 2005
|
|
