7ATJ
 
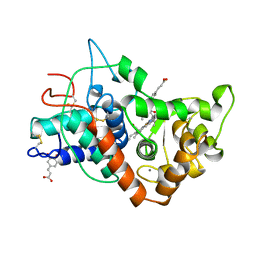 | | RECOMBINANT HORSERADISH PEROXIDASE C1A COMPLEX WITH CYANIDE AND FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, CYANIDE ION, ... | | Authors: | Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 1999-04-26 | | Release date: | 2000-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates.
J.Biol.Chem., 274, 1999
|
|
1M8R
 
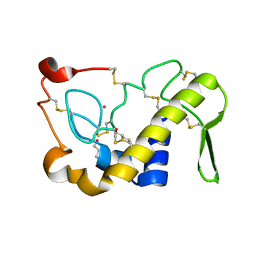 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 7.4) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase A2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
6GD9
 
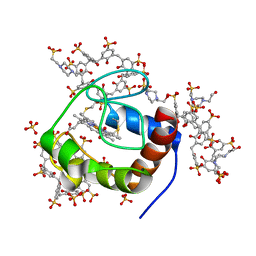 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P43212 form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytochrome c iso-1, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1ROS
 
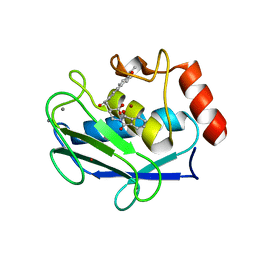 | | Crystal structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-[2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL)ETHYL]-4-(4'-ETHOXY-1,1'-BIPHENYL-4-YL)-4-OXOBUTANOIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J.Mol.Biol., 341, 2004
|
|
6GD6
 
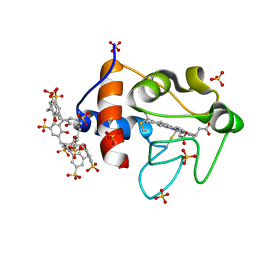 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with ammonium sulfate | | Descriptor: | Cytochrome c iso-1, HEME C, SULFATE ION, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5FXJ
 
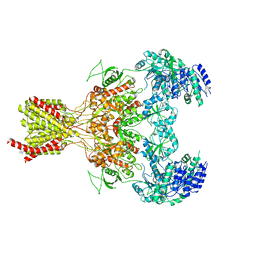 | | GluN1b-GluN2B NMDA receptor structure-Class X | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, NMDA 1, NMDA 2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa H, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
6V9W
 
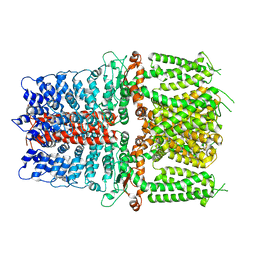 | | Structure of TRPA1 (ligand-free) with bound calcium, LMNG | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Zhao, J, Lin King, J.V, Paulsen, C.E, Cheng, Y, Julius, D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Irritant-evoked activation and calcium modulation of the TRPA1 receptor.
Nature, 585, 2020
|
|
6A5E
 
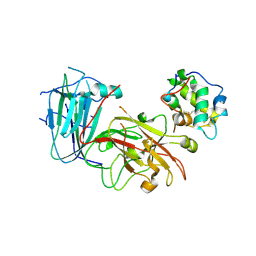 | | Crystal structure of plant peptide RALF23 in complex with FER and LLG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GPI-anchored protein LLG2, ... | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2018-06-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.766 Å) | | Cite: | Mechanisms of RALF peptide perception by a heterotypic receptor complex.
Nature, 572, 2019
|
|
6O77
 
 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, calcium-bound state | | Descriptor: | CALCIUM ION, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Diver, M.M, Cheng, Y, Julius, D. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
4AOE
 
 | | Biomphalaria glabrata Acetylcholine-binding protein type 2 (BgAChBP2) | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN TYPE 2 | | Authors: | Saur, M, Moeller, V, Kapetanopoulos, K, Braukmann, S, Gebauer, W, Tenzer, S, Markl, J. | | Deposit date: | 2012-03-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Acetylcholine-Binding Protein in the Hemolymph of the Planorbid Snail Biomphalaria Glabrata is a Pentagonal Dodecahedron (60 Subunits)
Plos One, 7, 2012
|
|
2FTB
 
 | | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to oleic acid | | Descriptor: | Fatty acid-binding protein 2, liver, OLEIC ACID | | Authors: | Capaldi, S, Guariento, M, Perduca, M, Di Pietro, S.M, Santome, J.A, Monaco, H.L. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of axolotl (Ambystoma mexicanum) liver bile acid-binding protein bound to cholic and oleic acid
Proteins, 64, 2006
|
|
4FFP
 
 | | PylC in complex with L-lysine-Ne-D-ornithine (cocrystallized with L-lysine and D-ornithine) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N~6~-D-ornithyl-L-lysine, ... | | Authors: | Quitterer, F, List, A, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of the 22nd genetically encoded amino acid pyrrolysine: structure and reaction mechanism of PylC at 1.5A resolution.
J.Mol.Biol., 424, 2012
|
|
9DHR
 
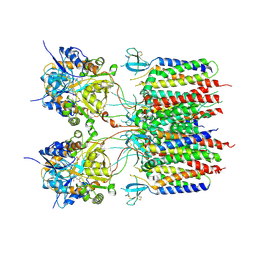 | | Glutamate activated state of the GluA2-gamma2 complex | | Descriptor: | GLUTAMIC ACID, Isoform Flip of Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Kumar Mondal, A, Carrillo, E, Jayaraman, V, Twomey, E.C. | | Deposit date: | 2024-09-04 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Glutamate gating of AMPA-subtype iGluRs at physiological temperatures.
Nature, 641, 2025
|
|
3BIG
 
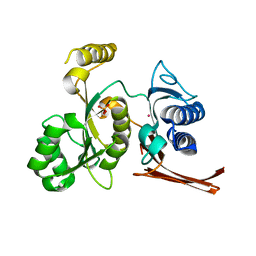 | | Crystal structure of the fructose-1,6-bisphosphatase GlpX from E.coli in complex with inorganic phosphate | | Descriptor: | Fructose-1,6-bisphosphatase class II glpX, PHOSPHATE ION, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A.F, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
9DHP
 
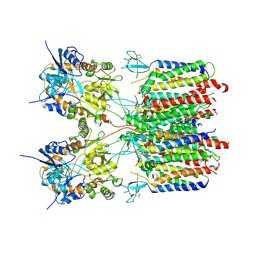 | | Resting state 1 of the GluA2-gamma2 complex | | Descriptor: | Isoform Flip of Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Kumar Mondal, A, Carrillo, E, Jayaraman, V, Twomey, E.C. | | Deposit date: | 2024-09-04 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Glutamate gating of AMPA-subtype iGluRs at physiological temperatures.
Nature, 641, 2025
|
|
4YYE
 
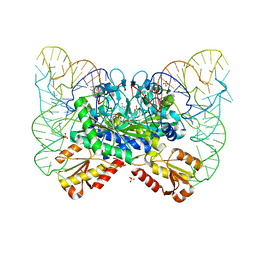 | |
7Z6X
 
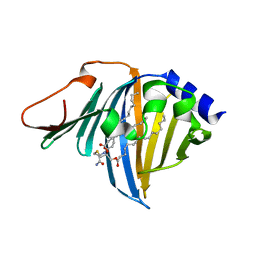 | | Complex of E. coli LolA R43L mutant and lipoprotein | | Descriptor: | Outer-membrane lipoprotein carrier protein, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Kaplan, E, Greene, N.P, Koronakis, V. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of lipoprotein recognition by the bacterial Lol trafficking chaperone LolA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6RTH
 
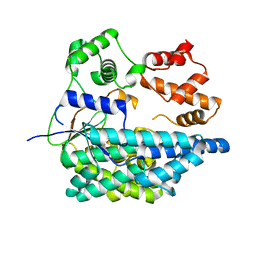 | |
3MEY
 
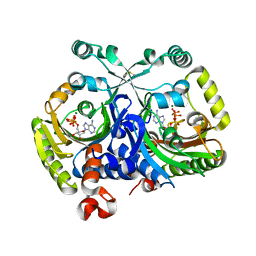 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bll0957 protein, MAGNESIUM ION, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7M3Z
 
 | | Structure of TIM-3 in complex with N-(4-(8-chloro-2-mehtyl-5-oxo-5,6-dihydro-[1,2,4]triazolo[1,5-c]quinazolin-9-yl)-3-methylphenyl)methanesulfonamdide (compound 35) | | Descriptor: | CALCIUM ION, Hepatitis A virus cellular receptor 2, N-{4-[(4S,10aP)-8-chloro-2-methyl-5-oxo-5,6-dihydro[1,2,4]triazolo[1,5-c]quinazolin-9-yl]-3-methylphenyl}methanesulfonamide | | Authors: | Rietz, T.A. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
J.Med.Chem., 64, 2021
|
|
7YFF
 
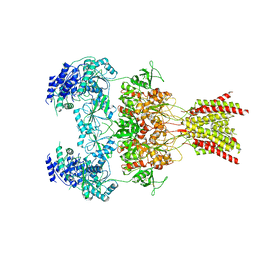 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
 | |
7YFR
 
 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5FXK
 
 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | Descriptor: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | Authors: | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | Deposit date: | 2016-03-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
