1L9B
 
 | | X-Ray Structure of the Cytochrome-c(2)-Photosynthetic Reaction Center Electron Transfer Complex from Rhodobacter sphaeroides in Type II Co-Crystals | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Okamura, M.Y, Yeh, A.P, Rees, D.C, Feher, G. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure determination of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides.
J.Mol.Biol., 319, 2002
|
|
1LE0
 
 | |
1TUI
 
 | | INTACT ELONGATION FACTOR TU IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Polekhina, G, Thirup, S, Kjeldgaard, M, Nissen, P, Lippmann, C, Nyborg, J. | | Deposit date: | 1996-05-23 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix unwinding in the effector region of elongation factor EF-Tu-GDP.
Structure, 4, 1996
|
|
3WNP
 
 | | D308A, F268V, D469Y, A513V, and Y515S quintuple mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoundecaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, R, Suzuki, N, Fujimoto, Z, Momma, M, Kimura, K, Kitamura, S, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular engineering of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040: structural determinants for the reaction product size and reactivity.
Biochem.J., 467, 2015
|
|
3W6S
 
 | | yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, MPR1 protein | | Authors: | Nasuno, R, Hirano, Y, Itoh, T, Hakoshima, T, Hibi, T, Takagi, H. | | Deposit date: | 2013-02-21 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of the yeast N-acetyltransferase Mpr1 involved in oxidative stress tolerance via proline metabolism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1L6R
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1LE3
 
 | |
1LAS
 
 | | Solution Structure of the B-DNA Duplex CGCGGTXTCCGCG (X=PdG) Containing the 1,N2-propanodeoxyguanosine Adduct with the Deoxyribose at C20 Opposite PdG in the C3' Endo Conformation. | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*(DNR)P*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*(P)P*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-29 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
1KKL
 
 | | L.casei HprK/P in complex with B.subtilis HPr | | Descriptor: | CALCIUM ION, HprK protein, PHOSPHOCARRIER PROTEIN HPR | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-12-10 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1QGK
 
 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1KTJ
 
 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
3WNK
 
 | | Crystal Structure of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | ACETATE ION, CADMIUM ION, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
1KXP
 
 | |
1LN2
 
 | | Crystal Structure of Human Phosphatidylcholine Transfer Protein in Complex with Dilinoleoylphosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
1R0E
 
 | | Glycogen synthase kinase-3 beta in complex with 3-indolyl-4-arylmaleimide inhibitor | | Descriptor: | 3-[3-(2,3-DIHYDROXY-PROPYLAMINO)-PHENYL]-4-(5-FLUORO-1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, CITRATE ANION, Glycogen synthase kinase-3 beta | | Authors: | Allard, J, Nikolcheva, T, Gong, L, Wang, J, Dunten, P, Avnur, Z, Waters, R, Sun, Q, Skinner, B. | | Deposit date: | 2003-09-20 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From genetics to therapeutics: the Wnt pathway and osteoporosis
To be Published
|
|
3WL9
 
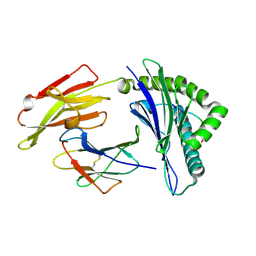 | | HLA-A24 in complex with HIV-1 Nef126-10(8I10F) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A, Han, C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Switching and emergence of CTL epitopes in HIV-1 infection
Retrovirology, 11, 2014
|
|
1LRA
 
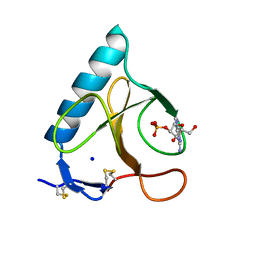 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
3WNN
 
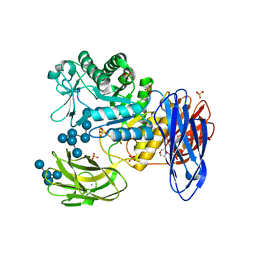 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WRP
 
 | |
1L8V
 
 | |
1LM4
 
 | | Structure of Peptide Deformylase from Staphylococcus aureus at 1.45 A | | Descriptor: | FE (III) ION, GLYCEROL, peptide deformylase PDF1 | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A. | | Deposit date: | 2002-04-30 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
1LME
 
 | | Crystal Structure of Peptide Deformylase from Thermotoga maritima | | Descriptor: | peptide deformylase | | Authors: | Kreusch, A, Spraggon, G, Lee, C.C, Klock, H, McMullan, D, Ng, K, Shin, T, Vincent, J, Warner, I, Ericson, C, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2002-05-01 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of peptide deformylases from streptococcus pneumoniae,staphylococcus aureus, thermotoga maritima, and pseudomonas aeruginosa: snapshots of the oxygen sensitivity of peptide deformylase
J.MOL.BIOL., 330, 2003
|
|
3ROW
 
 | | Crystal Structure of Xanthomonas campestri OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8487 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
1L9K
 
 | | dengue methyltransferase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Egloff, M.P, Benarroch, D, Selisko, B, Romette, J.L, Canard, B. | | Deposit date: | 2002-03-25 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An RNA cap (nucleoside-2'-O-) methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization
Embo J., 21, 2002
|
|
1LAQ
 
 | | Solution Structure of the B-DNA Duplex CGCGGTXTCCGCG (X=PdG) Containing the 1,N2-propanodeoxyguanosine Adduct with the Deoxyribose at C20 Opposite PdG in the C2' Endo Conformation. | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*(DNR)P*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*(P)P*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-29 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
