7MBM
 
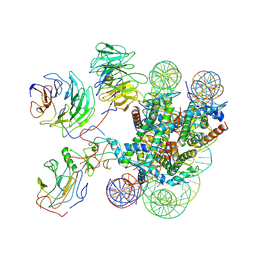 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode01 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
7A6D
 
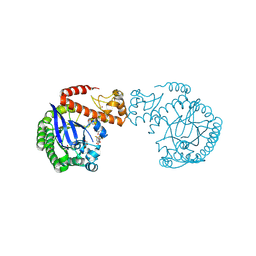 | | tRNA-guanine transglycosylase C158S/C281S/Y330C/H333A mutant in complex with rac-trans-4,4-difluorocyclopentane-1,2-diol | | Descriptor: | (1~{R},2~{R})-4,4-bis(fluoranyl)cyclopentane-1,2-diol, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Nguyen, D, Heine, A, Klebe, G. | | Deposit date: | 2020-08-25 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Targeting a Cryptic Pocket in a Protein-Protein Contact by Disulfide-Induced Rupture of a Homodimeric Interface.
Acs Chem.Biol., 16, 2021
|
|
7M4O
 
 | |
6PHO
 
 | |
6MDC
 
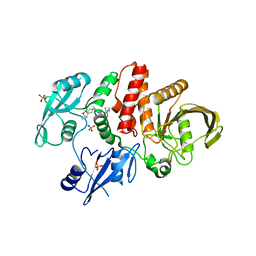 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor Pyrazolo-pyrimidinone 1 SHP389 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-[3-chloro-2-(cyclopropylamino)pyridin-4-yl]-5-methyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Fodor, M, Stams, T. | | Deposit date: | 2018-09-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of Fused Bicyclic Allosteric SHP2 Inhibitors.
J. Med. Chem., 62, 2019
|
|
8E5V
 
 | |
6ZU6
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6URK
 
 | | Barrier-to-autointegration factor soaked in Glycerol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6VGY
 
 | | 2.05 A resolution structure of MERS 3CL protease in complex with inhibitor 6b | | Descriptor: | N~2~-{[(trans-4-ethylcyclohexyl)oxy]carbonyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, Orf1a protein | | Authors: | Lovell, S, Battaile, K.P, Kashipathy, M.M, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6MDT
 
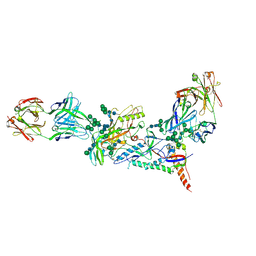 | | Crystal structure of the B41 SOSIP.664 Env trimer with PGT124 and 35O22 Fabs, in P63 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.816 Å) | | Cite: | Capturing the inherent structural dynamics of the HIV-1 envelope glycoprotein fusion peptide.
Nat Commun, 10, 2019
|
|
7MFY
 
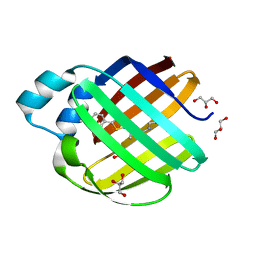 | |
6ZUB
 
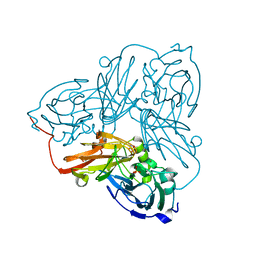 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 2 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
8EAA
 
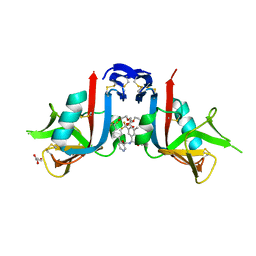 | | NKG2D complexed with inhibitor 4e | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-{(1S)-2-(dimethylamino)-1-[3-methyl-5-(trifluoromethyl)phenyl]-2-oxoethyl}-4-[4-(trifluoromethyl)phenyl]pyridine-3-carboxamide, ... | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6PHT
 
 | |
6PID
 
 | |
7A05
 
 | | NMR structure of D3-D4 domains of Vibrio vulnificus ribosomal protein S1 | | Descriptor: | 30S ribosomal protein S1 | | Authors: | Qureshi, N.S, Matzel, T, Cetiner, E.C, Schnieders, S, Jonker, H.R.A, Schwalbe, H, Fuertig, B. | | Deposit date: | 2020-08-06 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Vibrio vulnificus ribosomal protein S1 domains D3 and D4 provides insights into molecular recognition of single-stranded RNAs.
Nucleic Acids Res., 49, 2021
|
|
6ME5
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with agomelatine | | Descriptor: | OLEIC ACID, chimera protein of Melatonin receptor type 1A and GlgA glycogen synthase, ~{N}-[2-(7-methoxynaphthalen-1-yl)ethyl]ethanamide | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
7M5A
 
 | |
6VH7
 
 | | Doublet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
8DY3
 
 | | Crystal Structure of spFv GLK2 HL | | Descriptor: | SULFATE ION, spFv GLK2 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
7M4M
 
 | |
6ZUT
 
 | | Cu nitrite reductase MSOX series at 170K, dose point 5 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Anyonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6MEC
 
 | | Structure of a group II intron retroelement after DNA integration | | Descriptor: | MAGNESIUM ION, Maturase reverse transcriptase, SODIUM ION, ... | | Authors: | Haack, D, Yan, X, Zhang, C, Hingey, J, Lyumkis, D, Baker, T.S, Toor, N. | | Deposit date: | 2018-09-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures of a Group II Intron Reverse Splicing into DNA.
Cell, 178, 2019
|
|
8DY1
 
 | |
6PIL
 
 | | Antibody scFv-M204 monomeric state | | Descriptor: | CHLORIDE ION, scFv-M204 antibody | | Authors: | Abskharon, R, Sawaya, M.R, Seidler, P.M, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conformational antibody that binds tau oligomers and inhibits pathological seeding by extracts from donors with Alzheimer's disease.
J.Biol.Chem., 295, 2020
|
|
