8B3Z
 
 | |
5OR5
 
 | |
2O7G
 
 | |
1H3L
 
 | | N-terminal fragment of SigR from Streptomyces coelicolor | | Descriptor: | RNA POLYMERASE SIGMA FACTOR | | Authors: | Li, W, Stevenson, C.E.M, Burton, N, Jakimowicz, P, Paget, M.S.B, Buttner, M.J, Lawson, D.M, Kleanthous, C. | | Deposit date: | 2002-09-10 | | Release date: | 2002-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | Identification and Structure of the Anti-Sigma Factor-Binding Domain of the Disulfide-Stress Regulated Sigma Factor Sigma(R) from Streptomyces Coelicolor
J.Mol.Biol., 323, 2002
|
|
3MZY
 
 | |
4LUP
 
 | | Crystal structure of the complex formed by region of E. coli sigmaE bound to its -10 element non template strand | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor, region 2 of sigmaE of E. coli | | Authors: | Campagne, S, Marsh, M.E, Vorholt, J.A.V, Allain, F.H.-T, Capitani, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2Q1Z
 
 | |
4CXF
 
 | | Structure of CnrH in complex with the cytosolic domain of CnrY | | Descriptor: | CHLORIDE ION, CNRY, RNA POLYMERASE SIGMA FACTOR CNRH, ... | | Authors: | Maillard, A.P, Girard, E, Ziani, W, Petit-Hartlein, I, Kahn, R, Coves, J. | | Deposit date: | 2014-04-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of the Anti-Sigma Factor Cnry in Complex with the Sigma Factor Cnrh Shows a New Structural Class of Anti- Sigma Factors Targeting Extracytoplasmic-Function Sigma Factors.
J.Mol.Biol., 426, 2014
|
|
2Z2S
 
 | |
7QH5
 
 | |
4NQW
 
 | |
7LQ4
 
 | | Rr (RsiG)2-(c-di-GMP)2-WhiG complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), RsiG, WhiG | | Authors: | Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2021-02-12 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of a sigma-(c-di-GMP)-anti-sigma switch.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6DXO
 
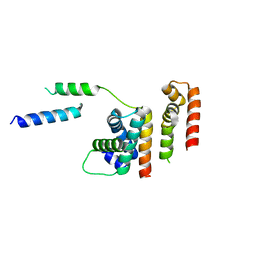 | | 1.8 A structure of RsbN-BldN complex. | | Descriptor: | BldN, RNA polymerase ECF-subfamily sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the RsbN-sigma BldN complex from Streptomyces venezuelae defines a new structural class of anti-sigma factor.
Nucleic Acids Res., 46, 2018
|
|
5UXX
 
 | |
5WUQ
 
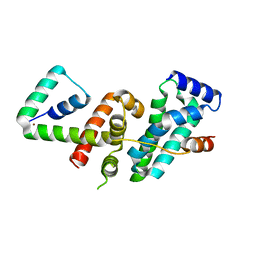 | | Crystal structure of SigW in complex with its anti-sigma RsiW, a zinc binding form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW, ZINC ION | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
5WUR
 
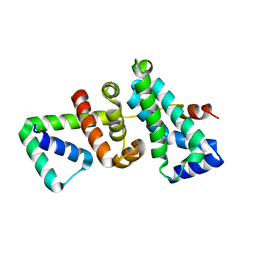 | | Crystal structure of SigW in complex with its anti-sigma RsiW, an oxdized form | | Descriptor: | Anti-sigma-W factor RsiW, ECF RNA polymerase sigma factor SigW | | Authors: | Devkota, S.R, Kwon, E, Ha, S.C, Kim, D.Y. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the regulation of Bacillus subtilis SigW activity by anti-sigma RsiW
PLoS ONE, 12, 2017
|
|
5XE7
 
 | |
8FTD
 
 | | Structure of Escherichia coli CedA in complex with transcription initiation complex | | Descriptor: | CHAPSO, Cell division activator CedA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, M, Vassyliev, N, Nudler, E. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-10 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | General transcription factor from Escherichia coli with a distinct mechanism of action.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6FBV
 
 | | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Das, K, Lin, W, Ebright, E. | | Deposit date: | 2017-12-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
8Z6G
 
 | |
5XJ0
 
 | | T. thermophilus RNA polymerase holoenzyme bound with gp39 and gp76 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ooi, W.Y, Murayama, Y, Mekler, V, Minakhin, L, Severinov, K, Yokoyama, S, Sekine, S. | | Deposit date: | 2017-04-28 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | A Thermus phage protein inhibits host RNA polymerase by preventing template DNA strand loading during open promoter complex formation
Nucleic Acids Res., 46, 2018
|
|
7RDQ
 
 | |
4MQ9
 
 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
9M84
 
 | |
