5MWW
 
 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
2K6X
 
 | | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1- Induced Compacted Structure | | Descriptor: | RNA polymerase sigma factor rpoD | | Authors: | Schwartz, E.C, Shekhtman, A, Dutta, K, Pratt, M.R, Cowburn, D, Darst, S, Muir, T.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Autoregulation of a Group 1 Bacterial Sigma Factor Involves the Formation of a Region 1.1 - Induced Compacted Structure
Chem.Biol., 15, 2008
|
|
7L7B
 
 | | Clostridioides difficile RNAP with fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Boyaci, H, Campbell, E.A, Darst, S.A, Chen, J. | | Deposit date: | 2020-12-28 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Basis of narrow-spectrum activity of fidaxomicin on Clostridioides difficile.
Nature, 604, 2022
|
|
8FTD
 
 | | Structure of Escherichia coli CedA in complex with transcription initiation complex | | Descriptor: | CHAPSO, Cell division activator CedA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Liu, M, Vassyliev, N, Nudler, E. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | General transcription factor from Escherichia coli with a distinct mechanism of action.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8AD1
 
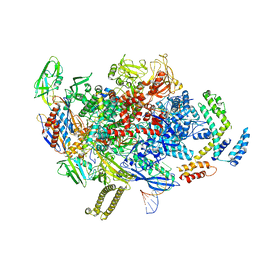 | |
6B6H
 
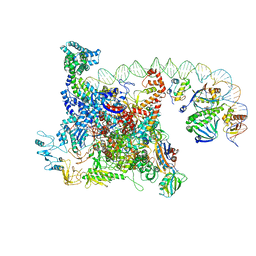 | | The cryo-EM structure of a bacterial class I transcription activation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
8X6F
 
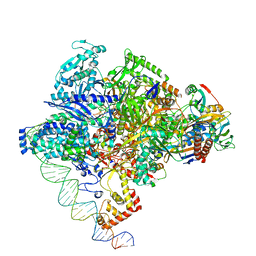 | | Cryo-EM structure of Staphylococcus aureus sigA-dependent RNAP-promoter open complex | | Descriptor: | DNA (71-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
8XA7
 
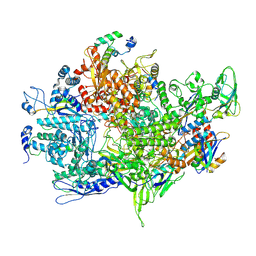 | | Cryo-EM structure of Bacillus subtilis RNAP,sigA and SPO1 gp33 complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Chen, M, Jin, Q, Yuan, S, Liu, B. | | Deposit date: | 2023-12-02 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | A phage encoded sigA-dependent transcription inhibitor also attacks host HelD-mediated defence system
To Be Published
|
|
7F75
 
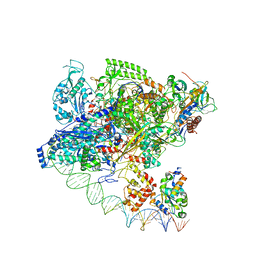 | | Cryo-EM structure of Spx-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of transcription activation by the global regulator Spx.
Nucleic Acids Res., 49, 2021
|
|
6WMT
 
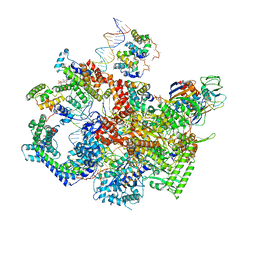 | | F. tularensis RNAPs70-(MglA-SspA)-ppGpp-PigR-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6VJS
 
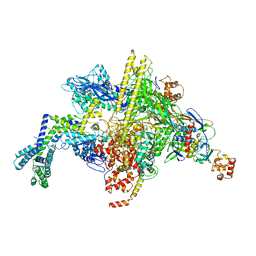 | | Escherichia coli RNA polymerase and ureidothiophene-2-carboxylic acid complex | | Descriptor: | 3-{[benzyl(ethyl)carbamoyl]amino}-5-(4-phenoxyphenyl)thiophene-2-carboxylic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murakami, K.S, Molodtsov, V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Evaluation of Bacterial RNA Polymerase Inhibitors in a Staphylococcus aureus -Based Wound Infection Model in SKH1 Mice.
Acs Infect Dis., 6, 2020
|
|
8Y6U
 
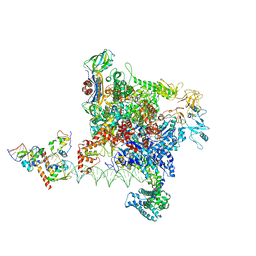 | |
7SZK
 
 | | Cryo-EM structure of 27a bound to E. coli RNAP and rrnBP1 promoter complex | | Descriptor: | (2S,7R,7aR,13aP,16Z,18E,20S,21S,22R,23R,24R,25S,26R,27S,28E)-5,21,23-trihydroxy-27-methoxy-2,4,16,20,22,24,26-heptamethyl-10-[4-(2-methylpropyl)piperazin-1-yl]-12-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1,6,15-trioxo-1,2,7,7a-tetrahydro-6H-2,7-(epoxypentadeca[1,11,13]trienoimino)[1]benzofuro[4,5-a]phenoxazin-25-yl acetate, DNA (5'-D(P*CP*TP*CP*GP*TP*AP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2021-11-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Optimization of Benzoxazinorifamycins to Improve Mycobacterium tuberculosis RNA Polymerase Inhibition and Treatment of Tuberculosis.
Acs Infect Dis., 8, 2022
|
|
7SZJ
 
 | |
7DY6
 
 | | A refined cryo-EM structure of an Escherichia coli RNAP-promoter open complex (RPo) with SspA | | Descriptor: | DNA (63-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lin, W. | | Deposit date: | 2021-01-20 | | Release date: | 2021-11-17 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | A unique binding between SspA and RNAP beta' NTH across low-GC Gram-negative bacteria facilitates SspA-mediated transcription regulation.
Biochem.Biophys.Res.Commun., 583, 2021
|
|
5UAG
 
 | | Escherichia coli RNA polymerase mutant - RpoB D516V | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.399 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
5UAQ
 
 | | Escherichia coli RNA polymerase RpoB H526Y mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
7CKQ
 
 | |
7C97
 
 | |
7CHW
 
 | |
6K4Y
 
 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
8JO2
 
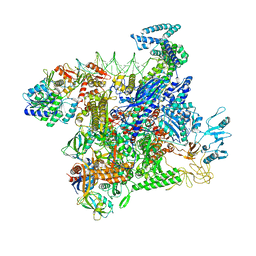 | | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA | | Descriptor: | DNA (65-MER), DNA-binding transcriptional regulator BasR, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lou, Y.-C, Huang, H.-Y, Chen, C, Wu, K.-P. | | Deposit date: | 2023-06-06 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis of transcriptional activation by the OmpR/PhoB-family response regulator PmrA.
Nucleic Acids Res., 51, 2023
|
|
7C17
 
 | |
8IGR
 
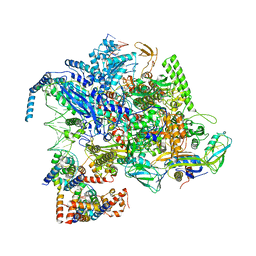 | | Cryo-EM structure of CII-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
