2JC7
 
 | |
6P56
 
 | |
8GPW
 
 | | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G | | Descriptor: | 1-[(~{Z})-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2~{S})-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxycyclopropane-1-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Song, D.Q, Li, Y.H. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G at 2.06 Angstroms resolution
To Be Published
|
|
3FV7
 
 | | OXA-24 beta-lactamase complex with SA4-44 inhibitor | | Descriptor: | (2S)-2-[[2-methanoyl-7-(methoxycarbonylamino)indolizin-3-yl]amino]-3-methyl-3-sulfino-butanoic acid, Beta-lactamase OXA-24 | | Authors: | Bou, G, Santillana, E, Sheri, A, Beceiro, A, Sampson, J.M, Kalp, M, Bethel, C.R, Distler, A.M, Drawz, S.M, Pagadala, S.R, Van den Akker, F, Bonomo, R.A, Romero, A, Buynak, J.D. | | Deposit date: | 2009-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase.
J.Am.Chem.Soc., 132, 2010
|
|
6B22
 
 | | Crystal structure OXA-24 beta-lactamase complexed with WCK 4234 by co-crystallization | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, Beta-lactamase, CHLORIDE ION | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
7DML
 
 | |
9MCZ
 
 | | Crystal structure of the transpeptidase domain of PBP2 from the Neisseria gonorrhoeae cephalosporin decreased susceptibility strain 35/02 in complex with boronate inhibitor VNRX-6884 | | Descriptor: | (3R)-3-{[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-(4-phosphonophenyl)acetyl]amino}-2-hydroxy-3,4-dihydro-2H-1,2-benzoxaborinine-8-carboxylic acid, Penicillin-binding protein 2 | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A new class of penicillin-binding protein inhibitors to address drug-resistant Neisseria gonorrhoeae.
Biorxiv, 2024
|
|
9MD0
 
 | | Crystal structure of the transpeptidase domain of PBP2 from the Neisseria gonorrhoeae cephalosporin decreased susceptibility strain 35/02 in complex with boronate inhibitor VNRX-6752 | | Descriptor: | (3R)-3-({(2R)-2-(4-carboxyphenyl)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]acetyl}amino)-2-hydroxy-3,4-dihydro-2H-1,2-benzoxaborinine-8-carboxylic acid, Penicillin-binding protein 2 | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | A new class of penicillin-binding protein inhibitors to address drug-resistant Neisseria gonorrhoeae.
Biorxiv, 2024
|
|
4YIN
 
 | | Crystal structure of the extended-spectrum beta-lactamase OXA-145 | | Descriptor: | Beta-lactamase, CITRATE ANION | | Authors: | Meziane-Cherif, D, Bonnet, R, Haouz, A, Courvalin, P. | | Deposit date: | 2015-03-02 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the loss of penicillinase and the gain of ceftazidimase activities by OXA-145 beta-lactamase in Pseudomonas aeruginosa.
J. Antimicrob. Chemother., 71, 2016
|
|
4MLL
 
 | | The 1.4 A structure of the class D beta-lactamase OXA-1 K70D complexed with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase OXA-1, ... | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2013-09-06 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural Origins of Oxacillinase Specificity in Class D beta-Lactamases.
Antimicrob.Agents Chemother., 58, 2014
|
|
2BG1
 
 | | Active site restructuring regulates ligand recognition in classA Penicillin-binding proteins (PBPs) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SULFATE ION | | Authors: | Macheboeuf, P, Di Guilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C5W
 
 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) ACYL-ENZYME COMPLEX (CEFOTAXIME) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | 1,2-ETHANEDIOL, CEFOTAXIME, C3' cleaved, ... | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-02 | | Release date: | 2005-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
2C6W
 
 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1A, ZINC ION | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-14 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
9D79
 
 | | OXA-58-NA-1-157 1.5 min complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, GLYCEROL | | Authors: | Smith, C.A, Maggiolo, A.O, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
9D7A
 
 | | OXA-58-NA-1-157 5 min complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Smith, C.A, Maggiolo, A.O, Toth, M, Vakulenko, S.B. | | Deposit date: | 2024-08-16 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decarboxylation of the Catalytic Lysine Residue by the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the OXA-58 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
6VBC
 
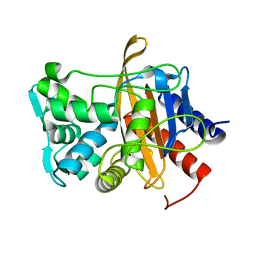 | |
6V6N
 
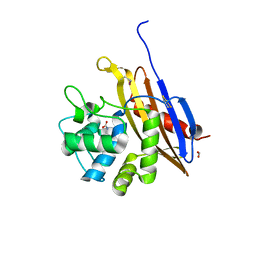 | | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens | | Descriptor: | Beta-lactamase, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of a class D beta-lactamase from Agrobacterium tumefaciens
To Be Published
|
|
6VBM
 
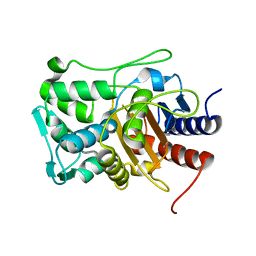 | | Crystal structure of a S310A mutant of PBP2 from Neisseria gonorrhoeae | | Descriptor: | PHOSPHATE ION, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Singh, A, Davies, C. | | Deposit date: | 2019-12-19 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mutations in Neisseria gonorrhoeae penicillin-binding protein 2 associated with extended-spectrum cephalosporin resistance create an energetic barrier against acylation via restriction of protein dynamics
J.Biol.Chem., 2020
|
|
8SQF
 
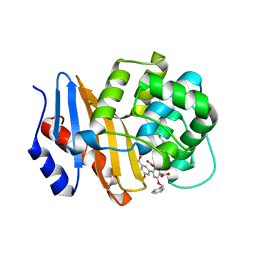 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQG
 
 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
4F94
 
 | | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Beta-lactamase, SULFATE ION | | Authors: | June, C.M, Vallier, B.C, Bonomo, R.A, Leonard, D.A, Powers, R.A. | | Deposit date: | 2012-05-18 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Class D Beta-Lactamase OXA-24 K84D in Acyl-Enzyme Complex with Oxacillin
To be Published
|
|
4S2J
 
 | | OXA-48 in complex with Avibactam at pH 6.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2M
 
 | | Crystal Structure of OXA-163 complexed with iodide in the active site | | Descriptor: | Beta-lactamase, IODIDE ION | | Authors: | Stojanoski, V, Hu, L, Palzkill, T.G, Prasad, B. | | Deposit date: | 2015-01-21 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Biochemistry, 54, 2015
|
|
6V1O
 
 | | Structure of OXA-48 bound to QPX7728 at 1.80 A | | Descriptor: | (1aR,7bS)-5-fluoro-2-hydroxy-1,1a,2,7b-tetrahydrocyclopropa[c][1,2]benzoxaborinine-4-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Boronic Acid QPX7728, an Ultrabroad-Spectrum Inhibitor of Serine and Metallo-beta-lactamases.
J.Med.Chem., 63, 2020
|
|
4S2O
 
 | | OXA-10 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, COBALT (II) ION | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
