8C3B
 
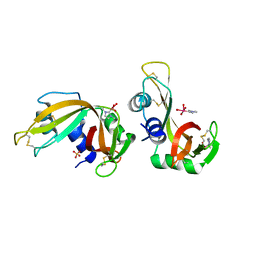 | | X-ray structure of RNase A upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | (1,3-dimethyl-2~{H}-imidazol-2-yl)-oxidanyl-oxidanylidene-ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tetrakis(oxidanyl)ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tris(oxidanyl)ruthenium, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Ruthenium(II)–Arene Complexes with Glycosylated NHC-Carbene Co-Ligands: Synthesis, Hydrolytic Behavior, and Binding to Biological Molecules
Organometallics, 42, 2023
|
|
5E5E
 
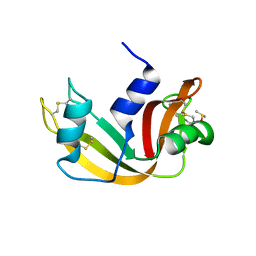 | |
9FYW
 
 | | X-ray structure of the adduct formed upon reaction of RNase A with [Ru2Cl(D-p-CNPhF)(O2CCH3)3] | | Descriptor: | 4-[11-(4-cyanophenyl)-1,5-bis($l^{1}-oxidanyl)-3,7-dimethyl-2,4,6,8-tetraoxa-9,11-diaza-1$l^{5},5$l^{5}-diruthenatricyclo[3.3.3.0^{1,5}]undecan-9-yl]benzenecarbonitrile, Ribonuclease pancreatic, SULFATE ION | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2024-07-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exchange of equatorial ligands in protein-bound paddlewheel Ru25+ complexes: new insights from X-ray crystallography and quantum chemistry
Inorg Chem Front, 11, 2024
|
|
7NPM
 
 | |
3A1R
 
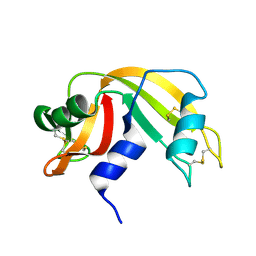 | |
6GOK
 
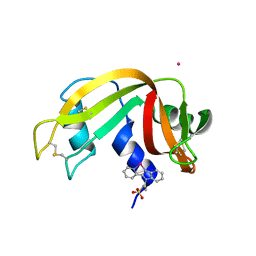 | | X-ray structure of the adduct formed upon reaction of bovine pancreatic ribonuclease with a Pd(II) complex bearing N,N-pyridylbenzimidazole derivative with an alkylated sulphonate side chain | | Descriptor: | N,N-pyridylbenzimidazole derivative-Pd complex, PALLADIUM ION, Ribonuclease pancreatic | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2018-06-01 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Exploring the interactions between model proteins and Pd(ii) or Pt(ii) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography.
Dalton Trans, 47, 2018
|
|
7TY1
 
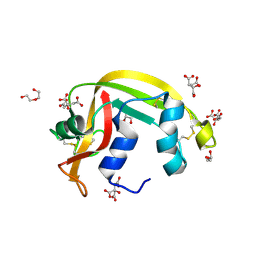 | | Crystal structure of apo eosinophil cationic protein (ribonuclease 3) from Macaca fascicularis (MfECP) | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, Eosinophil cationic protein, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
8AF0
 
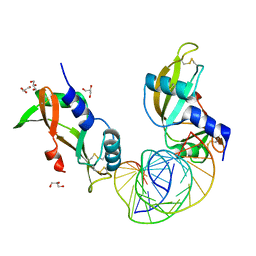 | | Crystal structure of human angiogenin and RNA duplex | | Descriptor: | Angiogenin, GLYCEROL, RNA (5'-R(*GP*CP*CP*CP*GP*CP*CP*UP*GP*UP*CP*AP*CP*GP*CP*GP*GP*GP*C)-3') | | Authors: | Sievers, K, Ficner, R. | | Deposit date: | 2022-07-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of angiogenin dimer bound to double-stranded RNA.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8F5X
 
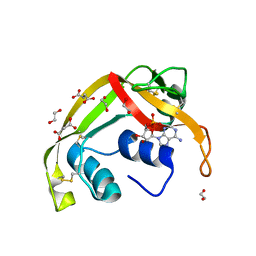 | | Crystal structure of human eosinophil-derived neurotoxin (EDN, ribonuclease 2) in complex with 5'-adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Non-secretory ribonuclease, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
4G8Y
 
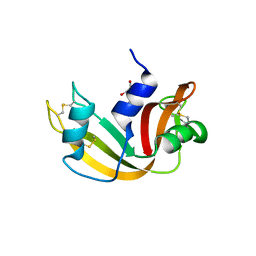 | | Crystal structure of Ribonuclease A in complex with 5b | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-methyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4G90
 
 | | Crystal structure of Ribonuclease A in complex with 5e | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-fluoro-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
1EIC
 
 | |
1EOW
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE A COMPLEXED WITH URIDYLYL(2',5')GUANOSINE (NON-PRODUCTIVE BINDING) | | Descriptor: | RIBONUCLEASE PANCREATIC, SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2000-03-24 | | Release date: | 2000-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Productive and nonproductive binding to ribonuclease A: X-ray structure of two complexes with uridylyl(2',5')guanosine.
Protein Sci., 9, 2000
|
|
5RSA
 
 | |
6RSA
 
 | |
8QEW
 
 | |
9JWH
 
 | | Crystal structure of native bovine pancreatic RNAs A | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Ahmad, M.S, Shah, N. | | Deposit date: | 2024-10-10 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyanide mediated conformational changes resulted in the displacement of sulfate ion from the active site of bovine pancreatic ribonuclease A.
Biochem.Biophys.Res.Commun., 736, 2024
|
|
9JWD
 
 | | Crystal structure of RNAs A treated with sodium cyanide | | Descriptor: | CHLORIDE ION, CYANIDE ION, GLYCEROL, ... | | Authors: | Ahmad, M.S, Shah, N. | | Deposit date: | 2024-10-10 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Cyanide mediated conformational changes resulted in the displacement of sulfate ion from the active site of bovine pancreatic ribonuclease A.
Biochem.Biophys.Res.Commun., 736, 2024
|
|
4G8V
 
 | | Crystal structure of Ribonuclease A in complex with 5a | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
6XVX
 
 | |
8R5V
 
 | | Crystal structure of bovine pancreatic ribonuclease A in complex with [Sp-PS]-mU-dT dinucleotide | | Descriptor: | 1-[(2~{R},4~{S},5~{R})-5-[[[(2~{R},6~{S})-2-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-6-(hydroxymethyl)morpholin-4-yl]-oxidanyl-phosphinothioyl]oxymethyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, DICHLORO-ACETIC ACID, Ribonuclease pancreatic | | Authors: | Dolot, R, Jastrzebska, K, Antonczyk, P. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | P-Stereodefined morpholino dinucleoside 3',5'-phosphorothioates.
Org.Biomol.Chem., 22, 2024
|
|
6XW0
 
 | |
5RAT
 
 | |
6MV6
 
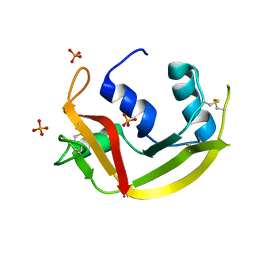 | | Crystal structure of RNAse 6 | | Descriptor: | PHOSPHATE ION, Ribonuclease K6 | | Authors: | Couture, J.-F, Doucet, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
7Z6G
 
 | |
