3QSV
 
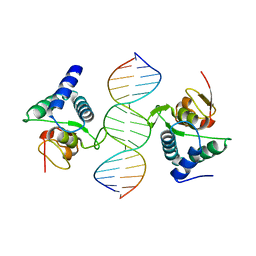 | |
1G88
 
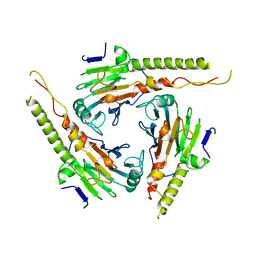 | | S4AFL3ARG515 MUTANT | | Descriptor: | SMAD4 | | Authors: | Chako, B.M, Qin, B, Lam, S.S, Correia, J.J, Lin, K. | | Deposit date: | 2000-11-16 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization.
Nat.Struct.Biol., 8, 2001
|
|
1DD1
 
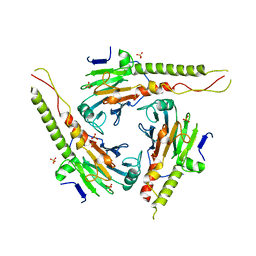 | |
1YGS
 
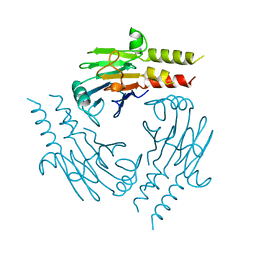 | | CRYSTAL STRUCTURE OF THE SMAD4 TUMOR SUPPRESSOR C-TERMINAL DOMAIN | | Descriptor: | SMAD4 | | Authors: | Shi, Y, Hata, A, Lo, R.S, Massague, J, Pavletich, N.P. | | Deposit date: | 1997-10-03 | | Release date: | 1998-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for mutational inactivation of the tumour suppressor Smad4.
Nature, 388, 1997
|
|
1U7V
 
 | | Crystal Structure of the phosphorylated Smad2/Smad4 heterotrimeric complex | | Descriptor: | Mothers against decapentaplegic homolog 2, Mothers against decapentaplegic homolog 4 | | Authors: | Chacko, B.M, Qin, B.Y, Tiwari, A, Shi, G, Lam, S, Hayward, L.J, de Caestecker, M, Lin, K. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of heteromeric smad protein assembly in tgf-Beta signaling
Mol.Cell, 15, 2004
|
|
5MEY
 
 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MEZ
 
 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MF0
 
 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5UWU
 
 | |
6YIC
 
 | |
5C4V
 
 | | Ski-like protein | | Descriptor: | GLYCEROL, Mothers against decapentaplegic homolog 4, NICKEL (II) ION, ... | | Authors: | Wallden, K, Nyman, T, Hallberg, B.M. | | Deposit date: | 2015-06-18 | | Release date: | 2016-10-12 | | Last modified: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SnoN Stabilizes the SMAD3/SMAD4 Protein Complex.
Sci Rep, 7, 2017
|
|
1MR1
 
 | | Crystal Structure of a Smad4-Ski Complex | | Descriptor: | Mothers against decapentaplegic homolog 4, Ski oncogene, ZINC ION | | Authors: | Wu, J.-W, Krawitz, A.R, Chai, J, Li, W, Zhang, F, Luo, K, Shi, Y. | | Deposit date: | 2002-09-17 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mechanism of Smad4 Recognition by the Nuclear Oncoprotein Ski: Insights on Ski-mediated Repression of TGF-beta Signaling
Cell(Cambridge,Mass.), 111, 2002
|
|
1U7F
 
 | | Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Mothers against decapentaplegic homolog 4 | | Authors: | Chacko, B.M, Qin, B.Y, Tiwari, A, Shi, G, Lam, S, Hayward, L.J, de Caestecker, M, Lin, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of heteromeric smad protein assembly in tgf-Beta signaling
Mol.Cell, 15, 2004
|
|
