5DOB
 
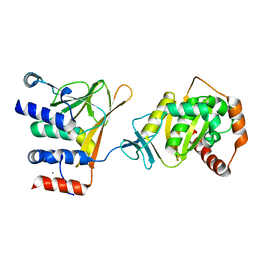 | | Crystal structure of the Human Cytomegalovirus Nuclear Egress Complex (NEC) | | Descriptor: | CALCIUM ION, Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ... | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
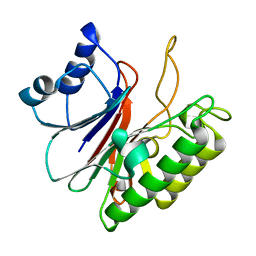
5EWT
 
 | |
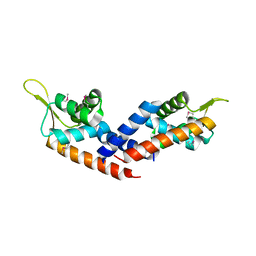
5HS5
 
 | |
7ZPN
 
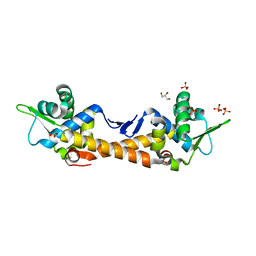 | | Crystal Structure of IscR from Dinoroseobacter shibae | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator, SULFATE ION | | Authors: | Lukat, P, Ploetzky, L, Blankenfeldt, W, Jahn, D, Haertig, E. | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dinoroseobacter shibae IscR homolog acts as a repressor for iron acquisition genes
To Be Published
|
|
5E3D
 
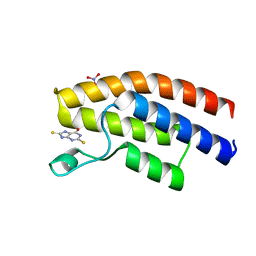 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED7 | | Descriptor: | 2,8-dithioxo-1,2,3,7,8,9-hexahydro-6H-purin-6-one, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-10-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
2ZOQ
 
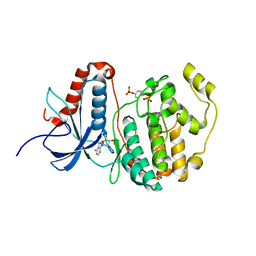 | | Structural dissection of human mitogen-activated kinase ERK1 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Mitogen-activated protein kinase 3, SODIUM ION, ... | | Authors: | Kinoshita, T, Tada, T, Nakae, S, Yoshida, I. | | Deposit date: | 2008-06-01 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human mono-phosphorylated ERK1 at Tyr204
Biochem.Biophys.Res.Commun., 377, 2008
|
|
8SOR
 
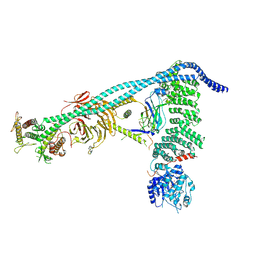 | | Structure of human PI3KC3-C1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beclin 1-associated autophagy-related key regulator, Beclin-1, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2023-04-29 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
8SBJ
 
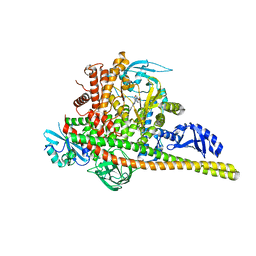 | | Co-structure Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform complexed with brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(1H-pyrazol-3-yl)-3H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
8SBC
 
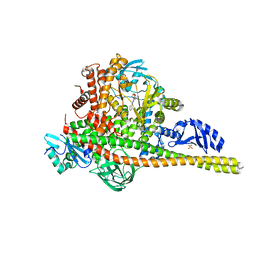 | | Co-structure of Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform and brain penetrant inhibitors | | Descriptor: | (2M)-7-[(3R)-3-methylmorpholin-4-yl]-5-[(3S)-3-methylmorpholin-4-yl]-2-(pyridin-2-yl)-1H-imidazo[4,5-b]pyridine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Knapp, M.S, Elling, R.A, Tang, J. | | Deposit date: | 2023-04-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Brain-Penetrant ATP-Competitive mTOR Inhibitors for CNS Syndromes.
J.Med.Chem., 66, 2023
|
|
8CYI
 
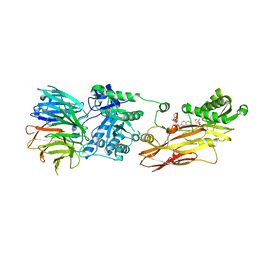 | | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, N-[(2-aminoquinolin-7-yl)methyl]-9-(2-hydroxyethyl)-2,3,4,9-tetrahydro-1H-carbazole-6-carboxamide, ... | | Authors: | Yadav, G.P, Wei, Z, Xiaozhi, Y, Chenglong, L, Jiang, Q. | | Deposit date: | 2022-05-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Commun Biol, 5, 2022
|
|
7TVA
 
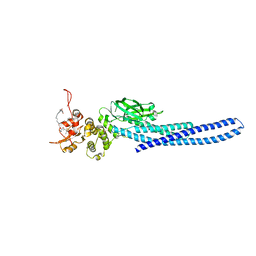 | | Stat5a Core in complex with AK2292 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVB
 
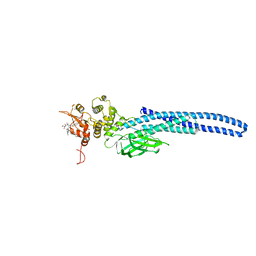 | | Stat5A Core in Complex with AK305 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
8JFV
 
 | | Crystal structure of Catabolite repressor acivator from E. coli in complex with sulisobenzone | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-oxidanyl-5-(phenylcarbonyl)benzenesulfonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Neetu, N, Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Sulisobenzone is a potent inhibitor of the global transcription factor Cra.
J.Struct.Biol., 215, 2023
|
|
2DS1
 
 | | Human cyclin dependent kinase 2 complexed with the CDK4 inhibitor | | Descriptor: | (13R,15S)-13-METHYL-16-OXA-8,9,12,22,24-PENTAAZAHEXACYCLO[15.6.2.16,9.1,12,15.0,2,7.0,21,25]HEPTACOSA-1(24),2,4,6,17(25 ),18,20-HEPTAENE-23,26-DIONE, Cell division protein kinase 2 | | Authors: | Ikuta, M. | | Deposit date: | 2006-06-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based drug design of a highly potent CDK1,2,4,6 inhibitor with novel macrocyclic quinoxalin-2-one structure
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8PO6
 
 | | Structure of Escherichia coli HrpA apo form | | Descriptor: | ATP-dependent RNA helicase HrpA, PHOSPHATE ION | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
2EFV
 
 | | Crystal Structure of a Hypothetical Protein(MJ0366) from Methanocaldococcus jannaschii | | Descriptor: | Hypothetical protein MJ0366, PHOSPHATE ION | | Authors: | Kumarevel, T.S, Karthe, P, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-26 | | Release date: | 2007-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure analysis of a hypothetical protein (MJ0366) from Methanocaldococcus jannaschii revealed a novel topological arrangement of the knot fold
Biochem. Biophys. Res. Commun., 482, 2017
|
|
2KA6
 
 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
8AVF
 
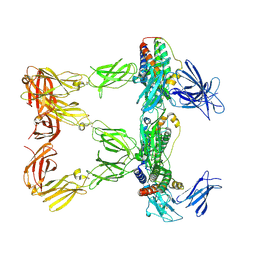 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (closed 3:3 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (6.45 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVO
 
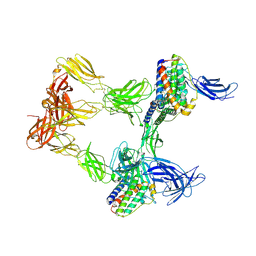 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (open 3:3 model). | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVE
 
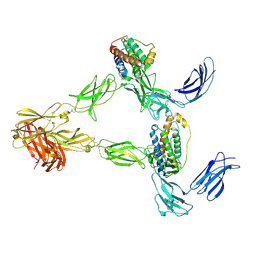 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (2:2 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.62 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2YDE
 
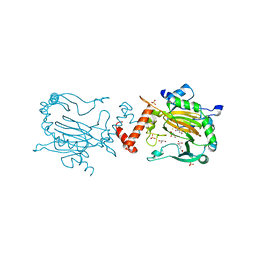 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH S-2-HYDROXYGLUTARATE | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Chowdhury, R, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2011-03-18 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
7DKH
 
 | | Crystal structure of the Ctr9/Paf1/Cdc73/Rtf1 quaternary complex | | Descriptor: | Cell division control protein 73, RNA polymerase II-associated protein 1, RNA polymerase-associated protein CTR9, ... | | Authors: | Chen, F.L, Liu, B.B, Guo, L, Li, D.F, Zhou, H, Long, J.F. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Core Module of the Yeast Paf1 Complex.
J.Mol.Biol., 434, 2021
|
|
6XU6
 
 | | Drosophila melanogaster Testis 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
5N63
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9c | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}4-[(4-fluorophenyl)methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
6XU8
 
 | | Drosophila melanogaster Ovary 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
