5NRR
 
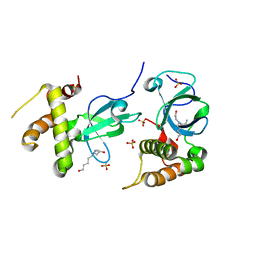 | |
8GBW
 
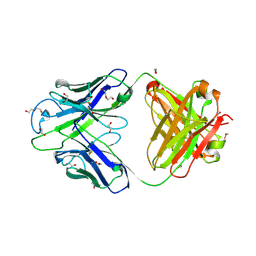 | | Crystal structure of PC39-23D, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PC39-23D Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
9HKZ
 
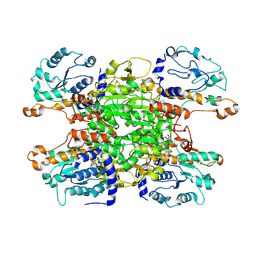 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 2 wide open, 1 open and 1 closed subunits
To Be Published
|
|
5ND6
 
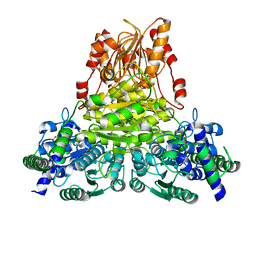 | | Crystal structure of apo transketolase from Chlamydomonas reinhardtii | | Descriptor: | 1,2-ETHANEDIOL, Transketolase | | Authors: | Fermani, S, Zaffagnini, M, Francia, F, Pasquini, M. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for the magnesium-dependent activation of transketolase from Chlamydomonas reinhardtii.
Biochim. Biophys. Acta, 1861, 2017
|
|
6CJK
 
 | | Anti HIV Fab 10A | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin Fab heavy chain, ... | | Authors: | Hangartner, L, Ward, A.B, Wilson, I.A, Oyen, D. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
8GBV
 
 | | Crystal structure of PC39-17A, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PC39-17A Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
5YWM
 
 | | Crystal structure of CK2a2 form-1 | | Descriptor: | Casein kinase II subunit alpha', NICOTINIC ACID, SULFATE ION | | Authors: | Tsuyuguchi, M, Kinoshita, T. | | Deposit date: | 2017-11-29 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Crystal structure of human CK2a2 in a new crystal form at 1.89 angstrom resolution
To Be Published
|
|
7TH1
 
 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B3 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(3-methylphenyl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
3KE4
 
 | |
6CEF
 
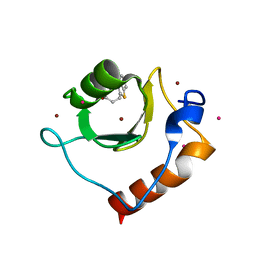 | | Crystal structure of fragment 3-(1,3-Benzothiazol-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(1,3-benzothiazol-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FYQ
 
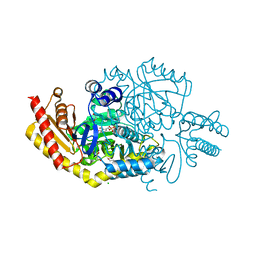 | |
7TGS
 
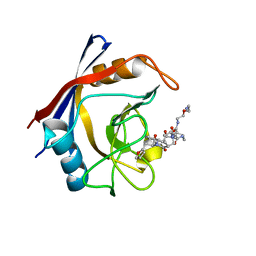 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor JOMBt | | Descriptor: | (4S,7R,11S,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7TGU
 
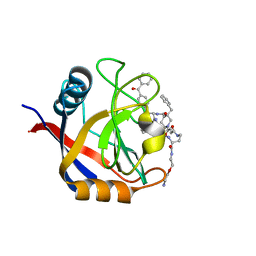 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor B1 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-4-[(4-benzoylphenyl)methyl]-7-benzyl-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Thakur, M.K, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
7CMB
 
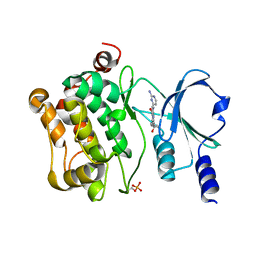 | | Crystal Structure of PAK4 in complex with inhibitor 41 | | Descriptor: | 1-(2-azanylpyrimidin-4-yl)-6-[2-(1-oxidanylcyclohexyl)ethynyl]indole-3-carboxamide, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2020-07-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Discovery of 6-ethynyl-1H-indole-3-carboxamide Derivatives as Highly Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors
to be published
|
|
6S51
 
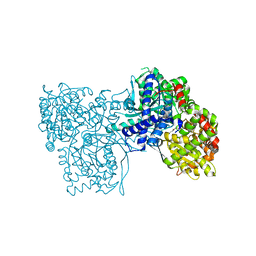 | | The crystal structure of glycogen phosphorylase in complex with 10 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-phenyl-1,3-thiazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
7TGT
 
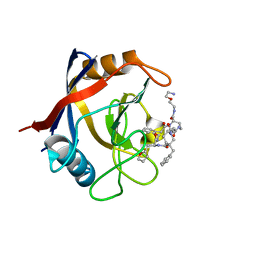 | | Structure of Cyclophilin D Peptidyl-Prolyl Isomerase Domain bound to Macrocyclic Inhibitor A26 | | Descriptor: | (4S,7S,11R,13E,19S)-N-[2-(2-aminoethoxy)ethyl]-7-benzyl-4-[(furan-2-yl)methyl]-3,6,12,15,21-pentaoxo-1,3,4,5,6,7,8,9,10,12,15,16,17,18,19,20,21,22-octadecahydro-2H-7,11-methano-2,5,11,16,20-benzopentaazacyclotetracosine-19-carboxamide, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Rangwala, A.M, Seeliger, M.A, Peterson, A.A, Liu, D.R. | | Deposit date: | 2022-01-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Discovery and molecular basis of subtype-selective cyclophilin inhibitors.
Nat.Chem.Biol., 18, 2022
|
|
6S4D
 
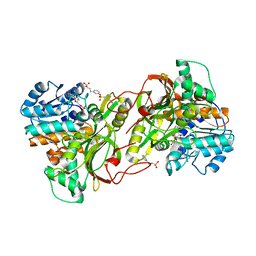 | |
5NMV
 
 | | Crystal structure of 2F22 Fab fragment against TFPI1 | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Svensson, L.A, Petersen, H.H. | | Deposit date: | 2017-04-07 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Factor Xa and VIIa inhibition by tissue factor pathway inhibitor is prevented by a monoclonal antibody to its Kunitz-1 domain.
J. Thromb. Haemost., 16, 2018
|
|
9EXX
 
 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 18. | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[1-methyl-5-(3-oxidanylidene-4~{H}-1,4-benzoxazin-7-yl)imidazol-4-yl]-~{N}-phenyl-benzamide, ETHANOL, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
6E4R
 
 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9B | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
2V6T
 
 | | Crystal structure of a complex of pterin-4a-carbinolamine dehydratase from Toxoplasma gondii with 7,8-dihydrobiopterin | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, PTERIN-4A-CARBINOLAMINE DEHYDRATASE | | Authors: | Cameron, S, Fyffe, S.A, Hunter, W.N. | | Deposit date: | 2007-07-20 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Toxoplasma Gondii Pterin-4A-Carbinolamine Dehydratase and Comparisons with Mammalian and Parasite Orthologues.
Mol.Biochem.Parasitol., 158, 2008
|
|
5NFJ
 
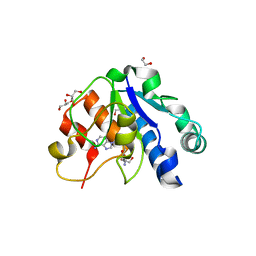 | | Crystal structure of the methyltransferase subunit of human mitochondrial Ribonuclease P (MRPP1) bound to S-adenosyl-methionine (SAM) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mitochondrial ribonuclease P protein 1, ... | | Authors: | Oerum, S, Kopec, J, Fitzpatrick, F, Newman, J.A, Chalk, R, Shrestha, L, Fairhead, M, Talon, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, C, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-14 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the methyltransferase subunit of human mitochondrial Ribonuclease P (MRPP1) bound to S-adenosyl-methionine (SAM)
To Be Published
|
|
8JHQ
 
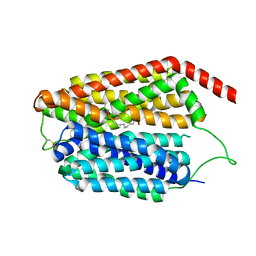 | | Cryo-EM structure of human S1P transporter SPNS2 bound with S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2,GlgA glycogen synthase | | Authors: | Pang, B, Yu, L.Y, Ren, R.B. | | Deposit date: | 2023-05-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis of Spns2-facilitated sphingosine-1-phosphate transport.
Cell Res., 34, 2024
|
|
6T4P
 
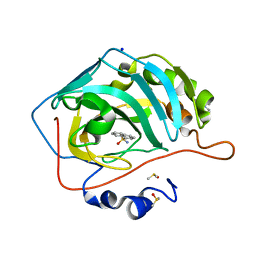 | | Human Carbonic anhydrase II bound by napthalene-1-sulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
8JNK
 
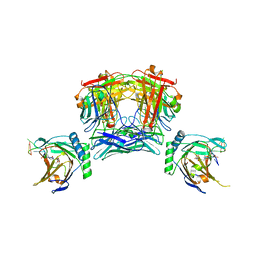 | |
