8AJV
 
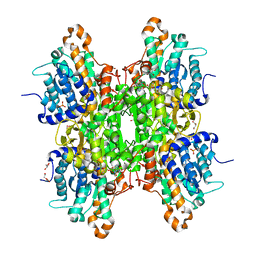 | | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa crystallized in the presence of K+ cations | | Descriptor: | Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa crystallized in the presence of K+ cations
To Be Published
|
|
8AJS
 
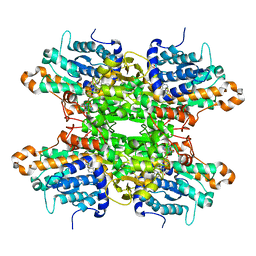 | | Crystal structure of the F324A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of the F324A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
8AJT
 
 | | Crystal structure of the H323A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of the H323A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
8AJU
 
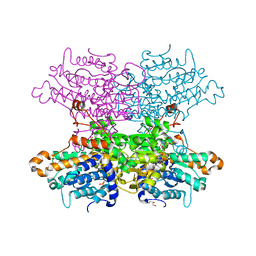 | | Crystal structure of the Q65A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Crystal structure of the Q65A mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
8AJW
 
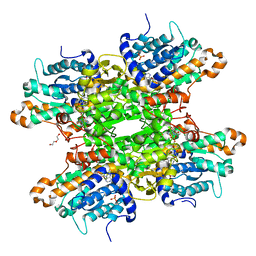 | | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Drozdzal, P, Wozniak, K, Malecki, P, Gawel, M, Komorowska, M, Brzezinski, K. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Crystal structure of the Q65N mutant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa cocrystallized with adenosine in the presence of K+ cations
To Be Published
|
|
8AZF
 
 | | Crystal structure of K449E variant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa in complex with adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Arning, A, Malecki, P, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of K449E variant of S-adenosyl-L-homocysteine hydrolase from Pseudomonas aeruginosa in complex with adenosine
To Be Published
|
|
7S3L
 
 | |
7ZD8
 
 | | Crystal structure of the R24E mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD7
 
 | | Crystal structure of the R24E/E352T double mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ZD9
 
 | | Crystal structure of the E352T mutant of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Wozniak, K, Brzezinski, K. | | Deposit date: | 2022-03-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
