4W5Z
 
 | |
4W5U
 
 | |
6SMK
 
 | | Crystal structure of catalytic domain A109H mutant of prophage-encoded M23 protein EnpA from Enterococcus faecalis. | | Descriptor: | Peptidase_M23 domain-containing protein, ZINC ION | | Authors: | Malecki, P.H, Mitkowski, P, Czapinska, H, Sabala, I. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Characterization of EnpA D,L-Endopeptidase from Enterococcus faecalis Prophage Provides Insights into Substrate Specificity of M23 Peptidases.
Int J Mol Sci, 22, 2021
|
|
3O5W
 
 | | Binding of kinetin in the active site of mistletoe lectin I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Malecki, P.H, Meyer, A, Rypniewski, W, Szymanski, M, Barciszewski, J, Betzel, C. | | Deposit date: | 2010-07-28 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of the plant hormone kinetin in the active site of Mistletoe Lectin I from Viscum album.
Biochim.Biophys.Acta, 1824, 2012
|
|
7O5L
 
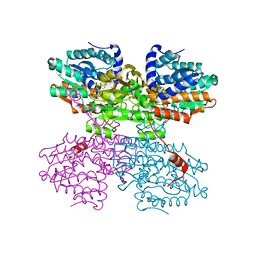 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Rb+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7O5M
 
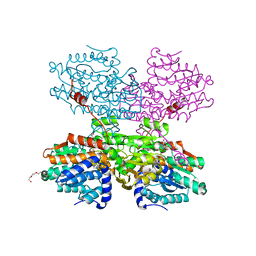 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803 cocrystallized with adenosine in the presence of Na+ cations | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Imiolczyk, B, Barciszewski, J, Czyrko-Horczak, J, Brzezinski, K. | | Deposit date: | 2021-04-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural insights into an unusual, alkali-metal-independent S-adenosyl-L-homocysteine hydrolase from Synechocystis sp. PCC 6803.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4MB4
 
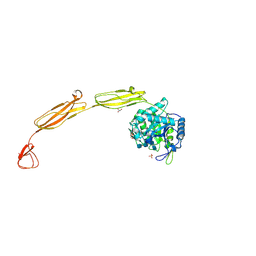 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB5
 
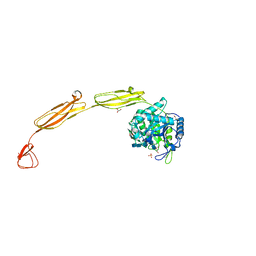 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4MB3
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella marina | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7BNH
 
 | |
7BNG
 
 | |
7BNI
 
 | |
4HMD
 
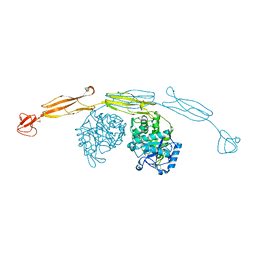 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HME
 
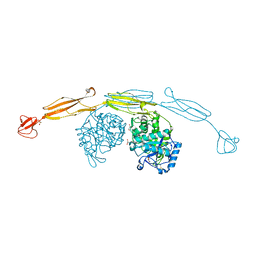 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HMC
 
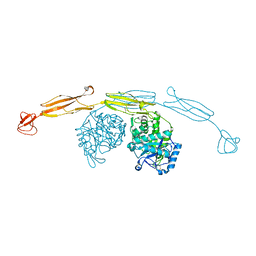 | | Crystal structure of cold-adapted chitinase from Moritella marina | | Descriptor: | Chitinase 60, GLYCEROL, SODIUM ION | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8CG1
 
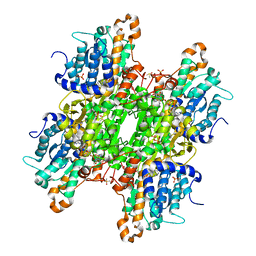 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment H12 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry H12
To be published
|
|
8CFY
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment H08 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry H08
To be published
|
|
8CFB
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment D02 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment D02
To be published
|
|
8CFL
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment E07 | | Descriptor: | 1-(1-methyl-1,2,3,4-tetrahydroquinolin-6-yl)methanamine, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry E07
To be published
|
|
8CFD
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment A07 | | Descriptor: | (2-aminopyridin-3-yl)methanol, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry A07
To be published
|
|
8CFN
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment F03 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry F03
To be published
|
|
8CFJ
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment D10 | | Descriptor: | 2-methyl-N-(4-methylphenyl)-L-alanine, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry D10
To be published
|
|
8CFR
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment G03 | | Descriptor: | 3-ethoxybenzene-1-carboximidamide, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry G03
To be published
|
|
8CFO
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment F04 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry F04
To be published
|
|
8CG0
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment H11 | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry H11
To be published
|
|
