4Y46
 
 | |
7YDX
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound RI-962 | | Descriptor: | 1-methyl-5-[2-(2-methylpropanoylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-phenylethyl]indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Wu, C, Luo, X, Wang, T, Lei, J, Yang, S. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor.
Nat Commun, 13, 2022
|
|
5T06
 
 | | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase YbgC, HEXANOYL-COENZYME A | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a putative acyl-CoA thioesterase EC709/ECK0725 from Escherichia coli in complex with Hexanoyl-CoA
To be published
|
|
9AUC
 
 | | Human Amylin1 Receptor in Complex with Gs and human Calcitonin Gene-Related Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin gene-related peptide 1, ... | | Authors: | Cao, J, Belousoff, M.J, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2024-02-28 | | Release date: | 2024-04-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structure of the Human Amylin 1 Receptor in Complex with CGRP and Gs Protein.
Biochemistry, 63, 2024
|
|
8EAQ
 
 | | Structure of the full-length IP3R1 channel determined at high Ca2+ | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 1, ... | | Authors: | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
9BS4
 
 | | DNA Ligase 1 E346A/E592A double mutant with 5'-rG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | KanalElamparithi, B, Caglayan, M. | | Deposit date: | 2024-05-12 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of LIG1 uncover the mechanism of sugar discrimination against 5'-RNA-DNA junctions during ribonucleotide excision repair.
J.Biol.Chem., 300, 2024
|
|
9BS3
 
 | | Wild type DNA Ligase 1 with 5'-rG:C | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | KanalElamparithi, B, Caglayan, M. | | Deposit date: | 2024-05-12 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structures of LIG1 uncover the mechanism of sugar discrimination against 5'-RNA-DNA junctions during ribonucleotide excision repair.
J.Biol.Chem., 300, 2024
|
|
5CSH
 
 | | Crystal Structure of CK2alpha with Compound 4 bound | | Descriptor: | 1-(2-chlorobiphenyl-4-yl)methanamine, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
9LB7
 
 | |
5WV1
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at 1.90 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Shokeen, A, Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with ribose sugar at
1.90 A resolution.
To Be Published
|
|
9AXM
 
 | | Crystal structure of ARAF/MEK1 complex with NST-628 and a RAF dimer | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 1, GLYCEROL, ... | | Authors: | Quade, B, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Pan-RAF-MEK Nondegrading Molecular Glue NST-628 Is a Potent and Brain-Penetrant Inhibitor of the RAS-MAPK Pathway with Activity across Diverse RAS- and RAF-Driven Cancers.
Cancer Discov, 14, 2024
|
|
7AGR
 
 | |
5MW3
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
7VOH
 
 | | The a-glucosidase QsGH13 from Qipengyuania seohaensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase QsGH13 | | Authors: | Huang, J, Zhai, X.Y. | | Deposit date: | 2021-10-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structure and Function Insight of the alpha-Glucosidase QsGH13 From Qipengyuania seohaensis sp. SW-135.
Front Microbiol, 13, 2022
|
|
7W7J
 
 | | Crystal structure of IrCp* immobilized apo-R52H-rHLFr (25 equiv) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Taher, M, Maity, B, Nakane, T, Abe, S, Mazumdar, S. | | Deposit date: | 2021-12-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Controlled Uptake of an Iridium Complex inside Engineered apo-Ferritin Nanocages: Study of Structure and Catalysis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8QPK
 
 | | Cryo-EM Structure of Pre-B+5'ss Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
6R2R
 
 | |
6R2N
 
 | | Crystal structure of KlGlk1 glucokinase from Kluyveromyces lactis | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Glucokinase-1 | | Authors: | Zak, K, Wator, E, Grudnik, P. | | Deposit date: | 2019-03-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Crystal Structure of Kluyveromyces lactis Glucokinase ( Kl Glk1).
Int J Mol Sci, 20, 2019
|
|
7AGP
 
 | | Structure of the AcylTransferase domain of Mycocerosic Acid Synthase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Mycocerosic acid synthase | | Authors: | Brison, Y, Nahoum, V, Mourey, L, Maveyraud, L. | | Deposit date: | 2020-09-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Extender Unit Specificity of Mycobacterial Polyketide Synthases.
Acs Chem.Biol., 15, 2020
|
|
8QP9
 
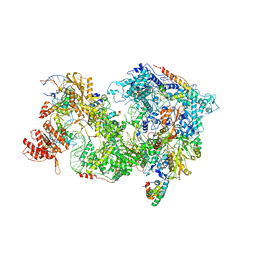 | | Cryo-EM Structure of Pre-B+AMPPNP Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Pre-mRNA-processing factor 6, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-30 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
7VJC
 
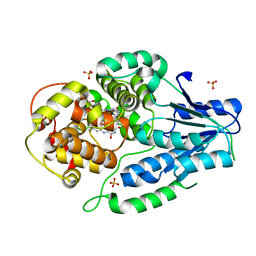 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (100 ns time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
6R2Z
 
 | |
7VJK
 
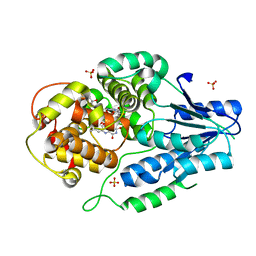 | | class II photolyase MmCPDII semiquinone to fully reduced TR-SFX studies (100 us time-point) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
2R9S
 
 | | c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 10, N-(tert-butyl)-4-[5-(pyridin-2-ylamino)quinolin-3-yl]benzenesulfonamide, ... | | Authors: | Habel, J. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5YGR
 
 | | Crystal structure of PLP bound Diaminopropionate ammonia lyase from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diaminopropionate ammonia lyase, ... | | Authors: | Geeta, D, Shveta, B, Shavithri, H.S, Murthy, M.R.N. | | Deposit date: | 2017-09-25 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparative structural and enzymatic studies on Salmonella typhimurium diaminopropionate ammonia lyase reveal its unique features
J. Struct. Biol., 202, 2018
|
|
