4DKU
 
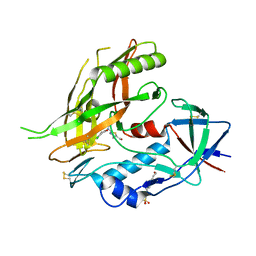 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with NBD-09027 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core, ... | | Authors: | Kwon, Y.D, Debnath, A.K, Kwong, P.D. | | Deposit date: | 2012-02-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4902 Å) | | Cite: | Binding mode characterization of NBD series CD4-mimetic HIV-1 entry inhibitors by X-ray structure and resistance study.
Antimicrob. Agents Chemother., 58, 2014
|
|
2O2H
 
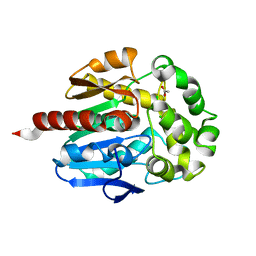 | | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane | | Descriptor: | 1,2-DICHLOROETHANE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of haloalkane dehalogenase Rv2579 from Mycobacterium tuberculosis complexed with 1,2-dichloroethane
TO BE PUBLISHED
|
|
4OCO
 
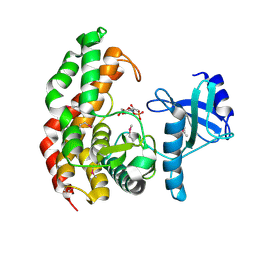 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6LJ8
 
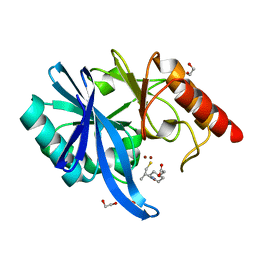 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04134 | | Descriptor: | 1,2-ETHANEDIOL, 2-[1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidin-4-yl]ethanoic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
1MER
 
 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP450 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
1MES
 
 | | HIV-1 MUTANT (I84V) PROTEASE COMPLEXED WITH DMP323 | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Ala, P, Chang, C.-H. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with cyclic urea inhibitors.
Biochemistry, 36, 1997
|
|
3NF6
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
2G81
 
 | | Crystal Structure of the Bowman-Birk Inhibitor from Vigna unguiculata Seeds in Complex with Beta-trypsin at 1.55 Angstrons Resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Bowman-Birk type seed trypsin and chymotrypsin inhibitor, ... | | Authors: | Freitas, S.M, Barbosa, J.A.R.G, Paulino, L.S, Teles, R.C.L, Esteves, G.F, Ventura, M.M. | | Deposit date: | 2006-03-01 | | Release date: | 2007-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Bowman-Birk Inhibitor from Vigna unguiculata Seeds in Complex with {beta}-Trypsin at 1.55 A Resolution and Its Structural Properties in Association with Proteinases
Biophys.J., 92, 2007
|
|
1HQD
 
 | | PSEUDOMONAS CEPACIA LIPASE COMPLEXED WITH TRANSITION STATE ANALOGUE OF 1-PHENOXY-2-ACETOXY BUTANE | | Descriptor: | (RP,SP)-O-(2R)-(1-PHENOXYBUT-2-YL)-METHYLPHOSPHONIC ACID CHLORIDE, CALCIUM ION, LIPASE | | Authors: | Luic, M, Tomic, S, Lescic, I, Ljubovic, E, Sepac, D, Sunjic, V, Vitale, L, Saenger, W, Kojic-Prodic, B. | | Deposit date: | 2000-12-15 | | Release date: | 2001-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex of Burkholderia cepacia lipase with transition state analogue of 1-phenoxy-2-acetoxybutane: biocatalytic, structural and modelling study.
Eur.J.Biochem., 268, 2001
|
|
4JAJ
 
 | | Crystal Structure of Aurora Kinase A in complex with BENZO[C][1,8]NAPHTHYRIDIN-6(5H)-ONE | | Descriptor: | Aurora kinase A, benzo[c][1,8]naphthyridin-6(5H)-one | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4E91
 
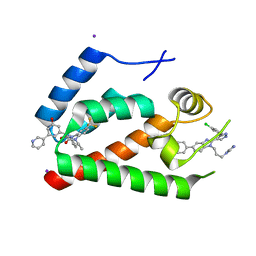 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BD3 | | Descriptor: | (3S)-1-ethyl-3-[3-hydroxy-5-(pyridin-3-yl)phenyl]-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct Effects of Two HIV-1 Capsid Assembly Inhibitor Families That Bind the Same Site within the N-Terminal Domain of the Viral CA Protein.
J.Virol., 86, 2012
|
|
3MXE
 
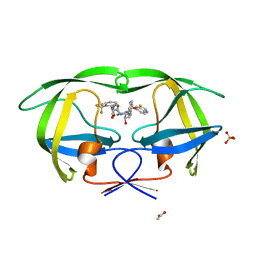 | | Crystal structure of HIV-1 protease inhibitor, KC32 complexed with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-2-oxo-3-[2-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
2AVV
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | Descriptor: | ACETIC ACID, CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
5GJV
 
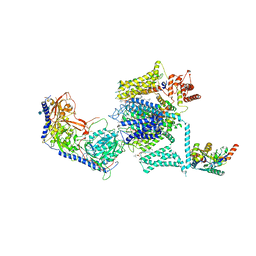 | | Structure of the mammalian voltage-gated calcium channel Cav1.1 complex at near atomic resolution | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Yan, Z, Li, Z.Q, Zhou, Q, Yan, N. | | Deposit date: | 2016-07-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the voltage-gated calcium channel Cav1.1 at 3.6 angstrom resolution
Nature, 537, 2016
|
|
4ENX
 
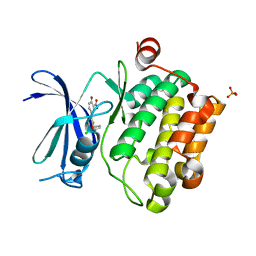 | | Crystal Structure of Pim-1 Kinase in complex with inhibitor (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-nitrobenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-nitrobenzylidene)-1,3-thiazolidin-4-one, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2BVY
 
 | | The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi | | Descriptor: | ACETATE ION, BETA-1,4-MANNANASE, CACODYLATE ION, ... | | Authors: | Le Nours, J, Anderson, L, Stoll, D, Stalbrand, H, Lo Leggio, L. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure and Characterization of a Modular Endo-Beta-1,4-Mannanase from Cellulomonas Fimi
Biochemistry, 44, 2005
|
|
3ZBX
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor 6-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl) quinoline. | | Descriptor: | 6-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J, Ryan, K. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
2ZBZ
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Shinkyo, R, Hayashi, K, Yoneda, S, Yamada, M, Kamakura, M, Ikushiro, S, Shiro, Y, Sakaki, T. | | Deposit date: | 2007-10-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of CYP105A1 (P450SU-1) in Complex with 1alpha,25-Dihydroxyvitamin D3
Biochemistry, 47, 2008
|
|
3IE3
 
 | | Structural basis for the binding of the anti-cancer compound 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) to human glutathione S-transferases | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]hexan-1-ol, GLUTATHIONE, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
4DWG
 
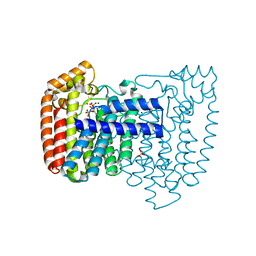 | | Crystal structure of Trypanosome cruzi farnesyl diphosphate synthase in complex with [2-(n-heptylamino)ethane-1,1-diyl]bisphosphonic acid and Mg2+ | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Farnesyl pyrophosphate synthase, ... | | Authors: | Aripirala, S, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design, synthesis, calorimetry, and crystallographic analysis of 2-alkylaminoethyl-1,1-bisphosphonates as inhibitors of Trypanosoma cruzi farnesyl diphosphate synthase.
J.Med.Chem., 55, 2012
|
|
2AID
 
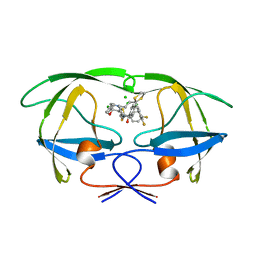 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
3NF9
 
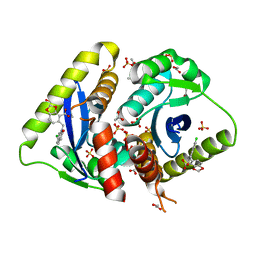 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(6-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
1Q7A
 
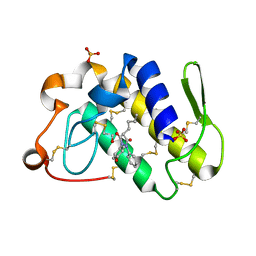 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
6BU7
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD130 1-[2-(Piperidin-4-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperidin-4-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-08 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
3NJ3
 
 | | Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Santos, C.R, Meza, A.N, Trindade, D.M, Ruller, R, Squina, F.M, Prade, R.A, Murakami, M.T. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
