5MZL
 
 | |
5N4O
 
 | |
5N4U
 
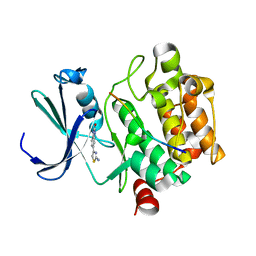 | | Crystal structure of human Pim-1 kinase in complex with a consensuspeptide and fragment like molekule 5-(2-amino-1,3-thiazol-4-yl)-1,3-dihydrobenzimidazol-2-one | | Descriptor: | 5-(2-azanyl-1,3-thiazol-4-yl)-1,3-dihydrobenzimidazol-2-one, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | A crystallographic fragment study with human Pim-1 kinase
to be published
|
|
5N5M
 
 | |
5N52
 
 | |
5N50
 
 | |
5N4R
 
 | |
5N4Z
 
 | |
6XTU
 
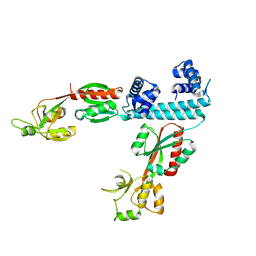 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM | | Descriptor: | Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6XTV
 
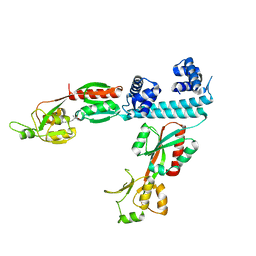 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND EFFECTOR ARG | | Descriptor: | ARGININE, Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
8FEX
 
 | |
5N4N
 
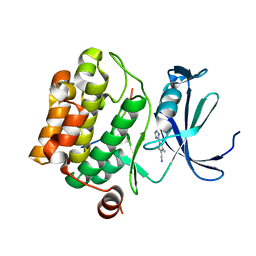 | | Crystal structure of human Pim-1 kinase in complex with a consensus peptide and fragment like molecule 3,4-dimethyl-5-(1H-1,2,4-triazol-3-yl)thiophene-2-carbonitrile | | Descriptor: | 3,4-dimethyl-5-(1~{H}-1,2,4-triazol-3-yl)thiophene-2-carbonitrile, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A crystallographic fragment study with human Pim-1 kinase
to be published
|
|
2O3F
 
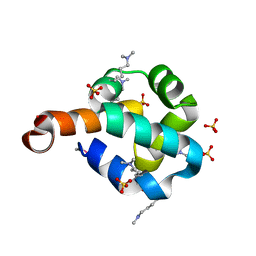 | | Structural Genomics, the crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168. | | Descriptor: | Putative HTH-type transcriptional regulator ybbH, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-01 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the N-terminal domain of the putative transcriptional regulator ybbH from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
6BH8
 
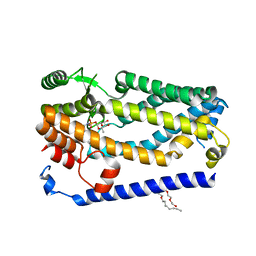 | | Crystal structure of ZMPSTE24 in complex with phosphoramidon | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CAAX prenyl protease 1 homolog, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Goblirsch, B.R, Arachea, B.T, Wiener, M.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Phosphoramidon inhibits the integral membrane protein zinc metalloprotease ZMPSTE24.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
3K1M
 
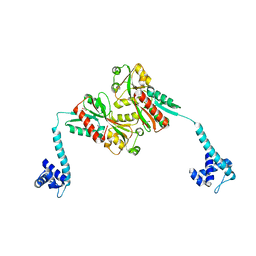 | | Crystal Structure of full-length BenM, R156H mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, HTH-type transcriptional regulator benM, ... | | Authors: | Ruangprasert, A, Momany, C, Neidle, E.L, Craven, S.H. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of Full-length BenM
To be Published
|
|
5O12
 
 | |
5N4X
 
 | |
5N51
 
 | |
5N4V
 
 | | Crystal structure of human Pim-1 kinase in complex with a consensuspeptide and fragment like molekule 2-cyclopropyl-4,5-dimethylthieno[5,4-d]pyrimidine-6-carboxylic acid | | Descriptor: | 2-cyclopropyl-4,5-dimethyl-thieno[2,3-d]pyrimidine-6-carboxylic acid, GLYCEROL, Pimtide, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A crystallographic fragment study with human Pim-1 kinase
to be published
|
|
5N4Y
 
 | | Crystal structure of human Pim-1 kinase in complex with a consensus peptide and fragment like molecule 2,5-dihydro-1H-isothiochromeno[3,4-d]pyrazol-3-one | | Descriptor: | 2,5-dihydro-1~{H}-isothiochromeno[4,3-c]pyrazol-3-one, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A crystallographic fragment study with human Pim-1 kinase
to be published
|
|
5N5L
 
 | |
5NDT
 
 | |
3K1P
 
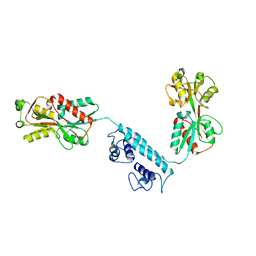 | |
5O11
 
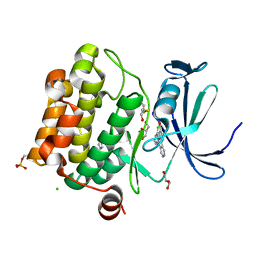 | |
6GXP
 
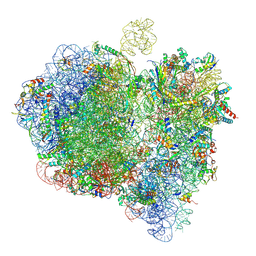 | | Cryo-EM structure of a rotated E. coli 70S ribosome in complex with RF3-GDPCP(RF3-only) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
