5IFR
 
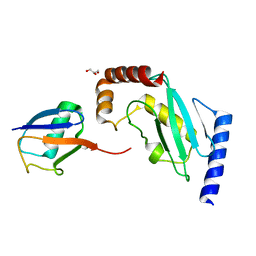 | | Structure of the stable UBE2D3-UbDha conjugate | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Pruneda, J.N, Mulder, M.P.C, Witting, K, Ovaa, H, Komander, D. | | Deposit date: | 2016-02-26 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cascading activity-based probe sequentially targets E1-E2-E3 ubiquitin enzymes.
Nat.Chem.Biol., 12, 2016
|
|
6OMB
 
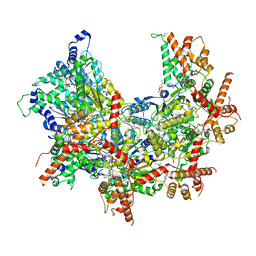 | | Cdc48 Hexamer (Subunits A to E) with substrate bound to the central pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
6AGG
 
 | |
6ZQD
 
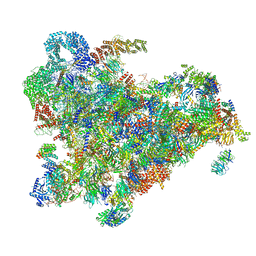 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6OPC
 
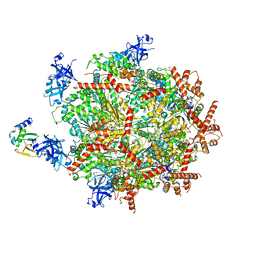 | | Cdc48 Hexamer in a complex with substrate and Shp1(Ubx Domain) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
1BUH
 
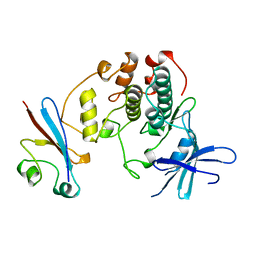 | |
3ILV
 
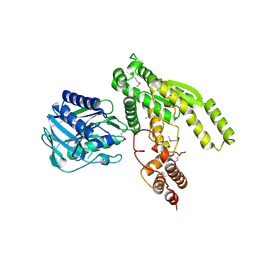 | |
6WJ3
 
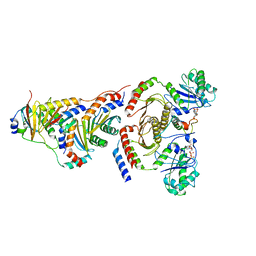 | |
6CES
 
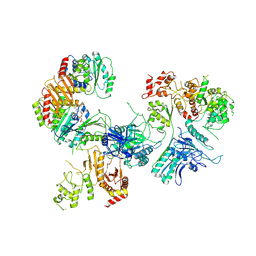 | | Cryo-EM structure of GATOR1-RAG | | Descriptor: | GATOR complex protein DEPDC5, GATOR complex protein NPRL2, GATOR complex protein NPRL3, ... | | Authors: | Shen, K, Huang, R.K, Brignole, E.J, Yu, Z, Sabatini, D.M. | | Deposit date: | 2018-02-12 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes.
Nature, 556, 2018
|
|
6WJ2
 
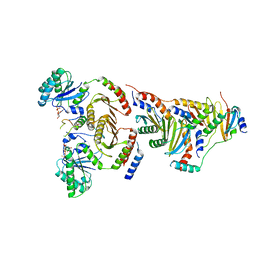 | | CryoEM structure of the SLC38A9-RagA-RagC-Ragulator complex in the pre-GAP state | | Descriptor: | 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(thiophosphonooxy)phosphoryl]oxy}phosphoryl]-alpha-L-arabinofuranosyl}-3,9-dihydro-1H-purine-2,6-dione, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fromm, S.A, Hurley, J.H. | | Deposit date: | 2020-04-11 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism for amino acid-dependent Rag GTPase nucleotide state switching by SLC38A9.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
2FNJ
 
 | |
6TH2
 
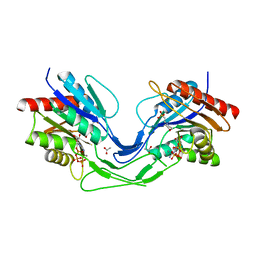 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
6THC
 
 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP and (4-hydroxyphenyl)(2,3,4-trihydroxyphenyl)methanone | | Descriptor: | (4-hydroxyphenyl)-[2,3,4-tris(oxidanyl)phenyl]methanone, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
3VBB
 
 | |
1TT5
 
 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3W31
 
 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
2NVU
 
 | | Structure of APPBP1-UBA3~NEDD8-NEDD8-MgATP-Ubc12(C111A), a trapped ubiquitin-like protein activation complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maltose binding protein/NEDD8-activating enzyme E1 catalytic subunit chimera, ... | | Authors: | Huang, D.T, Hunt, H.W, Zhuang, M, Ohi, M.D, Holton, J.M, Schulman, B.A. | | Deposit date: | 2006-11-13 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity.
Nature, 445, 2007
|
|
4JJQ
 
 | | Crystal structure of usp7-ntd with an e2 enzyme | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Saridakis, V. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1.
J. Biol. Chem., 288, 2013
|
|
6ZHS
 
 | | Uba1 bound to two E2 (Ubc13) molecules | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHT
 
 | | Uba1-Ubc13 disulfide mediated complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Schaefer, A, Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
4YC6
 
 | | CDK1/CKS1 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 1 | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
