4M2W
 
 | | Genetically engineered Carbonic Anhydrase IX in complex with Dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Pinard, M.P, Boone, C.D, Rife, B.D, Supuran, C.T, McKenna, R. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.
Bioorg.Med.Chem., 21, 2013
|
|
3KKV
 
 | | Structure of PKA with a protein Kinase B-selective inhibitor. | | Descriptor: | (2S)-1-{[6-furan-3-yl-5-(3-methyl-2H-indazol-5-yl)pyridin-3-yl]oxy}-3-(1H-indol-3-yl)propan-2-amine, PKI kinase inhibitor, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Elkins, P.A, Concha, N.O. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PKA with a protein Kinase B-selective inhibitor.
To be Published
|
|
1OIM
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 1-DEOXYNOJIRIMYCIN, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
3SDW
 
 | |
3T1A
 
 | |
3T3C
 
 | | Structure of HIV PR resistant patient derived mutant (comprising 22 mutations) in complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, BETA-MERCAPTOETHANOL, HIV-1 protease, ... | | Authors: | Rezacova, P, Kozisek, M, Konvalinka, J, Saskova, K.G. | | Deposit date: | 2011-07-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in HIV-1 gag and pol Compensate for the Loss of Viral Fitness Caused by a Highly Mutated Protease.
Antimicrob.Agents Chemother., 56, 2012
|
|
1OD0
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
2YK6
 
 | | Structure of Neisseria LOS-specific sialyltransferase (NST), in complex with CDP. | | Descriptor: | CMP-N-ACETYLNEURAMINATE-BETA-GALACTOSAMIDE-ALPHA-2,3-SIALYLTRANSFERASE, CYTIDINE-5'-DIPHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Lin, L.Y.C, Rakic, B, Chiu, C.P.C, Lameignere, E, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2011-05-25 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Mechanism of the Lipooligosaccharide Sialyltransferase from Neisseria Meningitidis
J.Biol.Chem., 286, 2011
|
|
2C3I
 
 | | CRYSTAL STRUCTURE OF HUMAN PIM1 IN COMPLEX WITH IMIDAZOPYRIDAZIN I | | Descriptor: | 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, PIMTIDE, PROTO-ONCOGENE SERINE THREONINE PROTEIN KINASE PIM1 | | Authors: | Philippakopoulos, P, Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K, Weigelt, J. | | Deposit date: | 2005-10-07 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis Identifies Imidazo[1,2- B]Pyridazines as Pim Kinase Inhibitors with in Vitro Antileukemic Activity.
Cancer Res., 67, 2007
|
|
2BFL
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH5 using 20mM ZnSO4 in buffer. 1mM DTT was used as a reducing agent. | | Descriptor: | AZIDE ION, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
1KLX
 
 | |
4FQV
 
 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H7 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQH
 
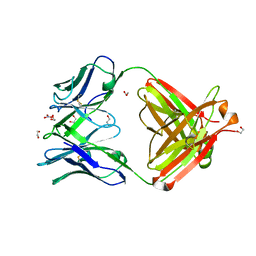 | | Crystal Structure of Fab CR9114 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, antibody CR9114 heavy chain, ... | | Authors: | Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
3T19
 
 | |
4R5N
 
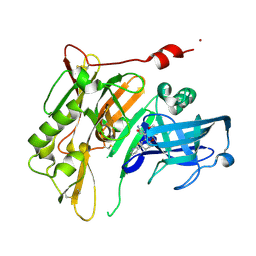 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4R,4a'S,10a'R)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2014-08-21 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
2GEE
 
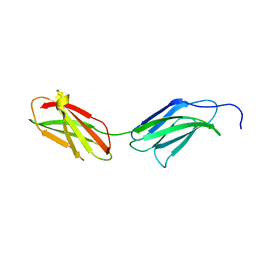 | | Crystal Structure of Human Type III Fibronectin Extradomain B and Domain 8 | | Descriptor: | hypothetical protein | | Authors: | Bencharit, S, Cui, C.B, Siddiqui, A, Howard-Williams, E.L, Aukhil, I. | | Deposit date: | 2006-03-19 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into fibronectin type III domain-mediated signaling.
J.Mol.Biol., 367, 2007
|
|
4FQY
 
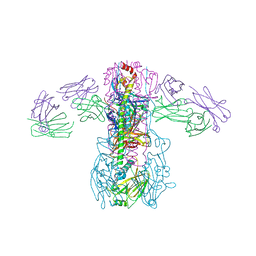 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H3 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.253 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQI
 
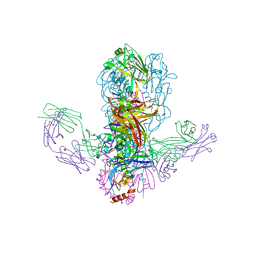 | |
3UUO
 
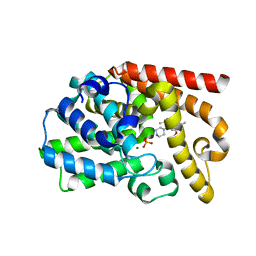 | | The discovery of potent, selectivity, and orally bioavailable pyrozoloquinolines as PDE10 inhibitors for the treatment of Schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(piperazin-1-yl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ho, G.D, Yang, S, Smotryski, J, Bercovici, A, Nechuta, T, Smith, E.M, McElroy, W, Tan, Z, Tulshian, D, Mckittrick, B, Greenlee, W.J, Hruza, A, Xiao, L, Rindgen, D, Guzzi, M, Zhang, X, Bleickardt, C, Mullins, D, Hodgson, R. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The discovery of potent, selective, and orally active pyrazoloquinolines as PDE10A inhibitors for the treatment of Schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3C3E
 
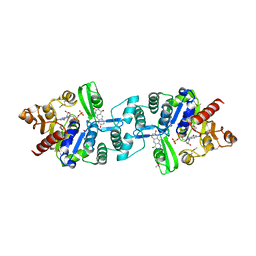 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and GDP. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
3SGW
 
 | |
1D4B
 
 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | Descriptor: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | Authors: | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | Deposit date: | 1999-10-02 | | Release date: | 1999-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
1ONJ
 
 | | Crystal structure of Atratoxin-b from Chinese cobra venom of Naja atra | | Descriptor: | Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Tu, X, Pan, G, Xu, C, Fan, R, Lu, W, Deng, W, Rao, P, Teng, M, Niu, L. | | Deposit date: | 2003-02-28 | | Release date: | 2004-02-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Purification, N-terminal sequencing, crystallization and preliminary structural determination of atratoxin-b, a short-chain alpha-neurotoxin from Naja atra venom.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2BVL
 
 | | Crystal structure of the catalytic domain of toxin B from Clostridium difficile in complex with UDP, Glc and manganese ion | | Descriptor: | HEXATANTALUM DODECABROMIDE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Reinert, D.J, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Function of Clostridium Difficile Toxin B.
J.Mol.Biol., 351, 2005
|
|
2BVM
 
 | | Crystal structure of the catalytic domain of toxin B from Clostridium difficile in complex with UDP, Glc and manganese ion | | Descriptor: | MANGANESE (II) ION, SULFATE ION, TOXIN B, ... | | Authors: | Reinert, D.J, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Function of Clostridium Difficile Toxin B.
J.Mol.Biol., 351, 2005
|
|
