6FWR
 
 | | Structure of DinG in complex with ssDNA | | Descriptor: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
4TYY
 
 | | DEAD-box helicase Mss116 bound to ssRNA and CDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TYW
 
 | | DEAD-box helicase Mss116 bound to ssRNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TZ6
 
 | | DEAD-box helicase Mss116 bound to ssRNA and UDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4U4C
 
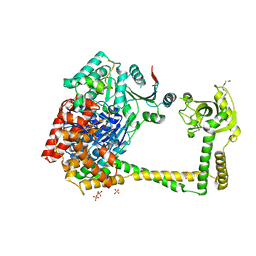 | | The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DOB1, CHLORIDE ION, ... | | Authors: | Falk, S, Weir, J.R, Hentschel, J, Reichelt, P, Bonneau, F, Conti, E. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Mol.Cell, 55, 2014
|
|
6HYU
 
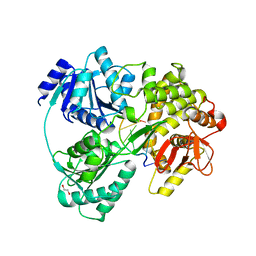 | | Crystal structure of DHX8 helicase bound to single stranded poly-adenine RNA | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DHX8, RNA (5'-R(*A*AP*A)-3'), ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
7UJB
 
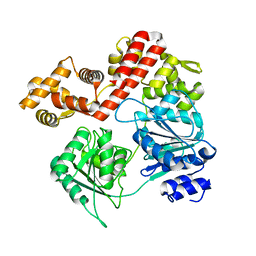 | | N-terminal domain deletion variant of Eta | | Descriptor: | DEAD/DEAH box RNA helicase | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2022-03-30 | | Release date: | 2022-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.12 Å) | | Cite: | The structure and activities of the archaeal transcription termination factor Eta detail vulnerabilities of the transcription elongation complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6UP4
 
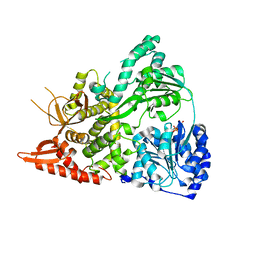 | |
4TYN
 
 | | DEAD-box helicase Mss116 bound to ssDNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.959 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
6HEG
 
 | | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpB, PHOSPHATE ION, ... | | Authors: | Xin, B.G, Chen, W.F, Rety, S, Dai, Y.X, Xi, X.G. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | Crystal structure of Escherichia coli DEAH/RHA helicase HrpB.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
4TMU
 
 | |
6UV1
 
 | | Crystal structure of RNA helicase DDX17 in complex of rU10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6HYT
 
 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.3 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
6UV2
 
 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
4U7D
 
 | | Structure of human RECQ-like helicase in complex with an oligonucleotide | | Descriptor: | ATP-dependent DNA helicase Q1, DNA oligonucleotide, ZINC ION | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3FHT
 
 | |
3FMP
 
 | | Crystal structure of the nucleoporin Nup214 in complex with the DEAD-box helicase Ddx19 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B, Nuclear pore complex protein Nup214 | | Authors: | Napetschnig, J, Debler, E.W, Blobel, G, Hoelz, A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FHO
 
 | | Structure of S. pombe Dbp5 | | Descriptor: | ATP-dependent RNA helicase dbp5 | | Authors: | Cheng, Z, Song, H. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides
J.Mol.Biol., 388, 2009
|
|
3G0H
 
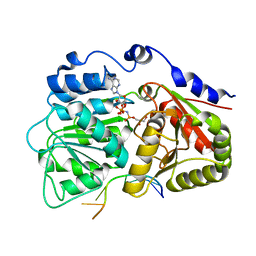 | | Human dead-box RNA helicase DDX19, in complex with an ATP-analogue and RNA | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase DDX19B, GLYCEROL, ... | | Authors: | Karlberg, T, Lehtio, L, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-28 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The DEXD/H-box RNA Helicase DDX19 Is Regulated by an {alpha}-Helical Switch.
J.Biol.Chem., 284, 2009
|
|
3EWS
 
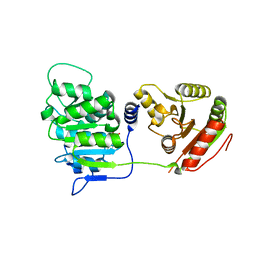 | | Human DEAD-box RNA-helicase DDX19 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B | | Authors: | Lehtio, l, Karlberg, t, Andersson, j, Arrowsmith, c.h, Berglund, h, Bountra, c, Collins, r, Dahlgren, l.g, Edwards, a.m, Flodin, s, Flores, a, Graslund, s, Hammarstrom, m, Johansson, a, Johansson, i, Kotenyova, t, Moche, m, Nilsson, m.e, Nordlund, p, Nyman, t, Olesen, k, Persson, c, Sagemark, j, Thorsell, a.g, Tresaugues, l, Van den berg, s, Weigelt, j, Welin, m, Wikstrom, m, Wisniewska, m, Schueler, h, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The DEXD/H-box RNA Helicase DDX19 Is Regulated by an {alpha}-Helical Switch.
J.Biol.Chem., 284, 2009
|
|
7AUD
 
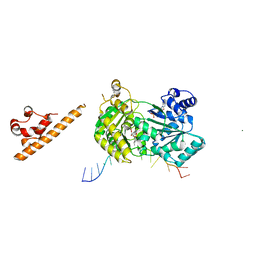 | |
7AUC
 
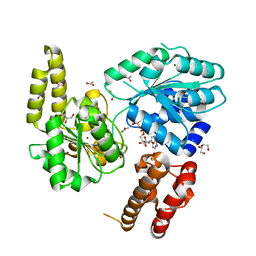 | |
7AQO
 
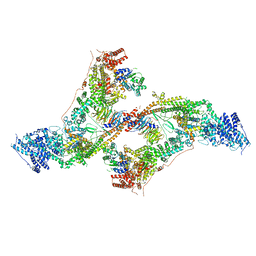 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7APX
 
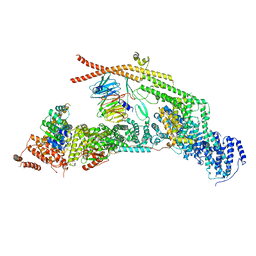 | | yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2,Tho2, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7APK
 
 | | Structure of the human THO - UAP56 complex | | Descriptor: | Spliceosome RNA helicase DDX39B, THO complex subunit 1, THO complex subunit 2, ... | | Authors: | Hohmann, U, Puehringer, T, Plaschka, C. | | Deposit date: | 2020-10-17 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human core transcription-export complex reveals a hub for multivalent interactions.
Elife, 9, 2020
|
|
