1PI5
 
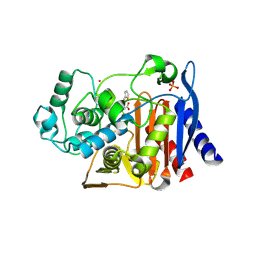 | | Structure of N289A mutant of AmpC in complex with SM2, carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Roth, T.A, Minasov, G, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2003-05-29 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Thermodynamic cycle analysis and inhibitor design against beta-lactamase.
Biochemistry, 42, 2003
|
|
1PI4
 
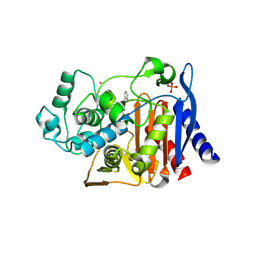 | | Structure of N289A mutant of AmpC in complex with SM3, a phenylglyclboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Roth, T.A, Minasov, G, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2003-05-29 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Thermodynamic cycle analysis and inhibitor design against beta-lactamase.
Biochemistry, 42, 2003
|
|
4OLG
 
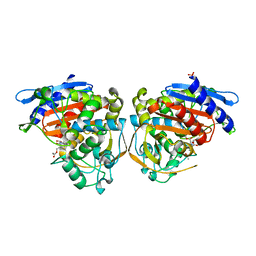 | | Crystal structure of AmpC beta-lactamase in complex with covalently bound N-formyl 7-aminocephalosporanic acid | | Descriptor: | (2R,5Z)-5-[(acetyloxy)methylidene]-2-[(1R)-1-(formylamino)-2-oxoethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Shoichet, B.K, Barelier, S. | | Deposit date: | 2014-01-23 | | Release date: | 2014-05-28 | | Last modified: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Substrate deconstruction and the nonadditivity of enzyme recognition.
J.Am.Chem.Soc., 136, 2014
|
|
3SH8
 
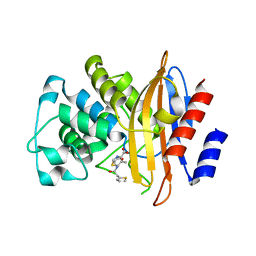 | | Crystal structure of fluorophore-labeled beta-lactamase PenP in complex with cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Wong, W.-T, Zhao, Y.-X, Leung, Y.-C. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Increased structural flexibility at the active site of a fluorophore-conjugated beta-lactamase distinctively impacts its binding toward diverse cephalosporin antibiotics
J.Biol.Chem., 286, 2011
|
|
4OLD
 
 | | Crystal structure of AmpC beta-lactamase in complex with the product form of (6R,7R)-7-amino-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid | | Descriptor: | (2R)-2-[(R)-amino(carboxy)methyl]-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Shoichet, B.K, Barelier, S. | | Deposit date: | 2014-01-23 | | Release date: | 2014-05-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Substrate deconstruction and the nonadditivity of enzyme recognition.
J.Am.Chem.Soc., 136, 2014
|
|
4OOY
 
 | | Avibactam and class C beta-lactamases: mechanism of inhibition, conservation of binding pocket and implications for resistance | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Lahiri, S.D, Olivier, N.B, Alm, R.A. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-20 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Avibactam and Class C beta-Lactamases: Mechanism of Inhibition, Conservation of the Binding Pocket, and Implications for Resistance.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NET
 
 | | Crystal structure of ADC-1 beta-lactamase | | Descriptor: | AmpC, GLYCEROL, NITRATE ION | | Authors: | Bhattacharya, M, Toth, M, Antunes, N.T, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase ADC-1 from Acinetobacter baumannii.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NK3
 
 | | Amp-c beta-lactamase (pseudomonas aeruginosa) in complex with mk-7655 | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-7655, a beta-lactamase inhibitor for combination with Primaxin().
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3SOI
 
 | | Crystallographic structure of Bacillus licheniformis beta-lactamase W210F/W229F/W251F at 1.73 angstrom resolution | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Acierno, J.P, Capaldi, S, Risso, V.A, Monaco, H.L, Ermacora, M.R. | | Deposit date: | 2011-06-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | X-ray evidence of a native state with increased compactness populated by tryptophan-less B. licheniformis beta-lactamase.
Protein Sci., 21, 2012
|
|
3SH7
 
 | | Crystal structure of fluorophore-labeled beta-lactamase PenP | | Descriptor: | 1-[6-(dimethylamino)naphthalen-2-yl]ethanone, Beta-lactamase | | Authors: | Wong, W.-T, Zhao, Y.-X, Leung, Y.-C. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Increased structural flexibility at the active site of a fluorophore-conjugated beta-lactamase distinctively impacts its binding toward diverse cephalosporin antibiotics
J.Biol.Chem., 286, 2011
|
|
4N9K
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
3SH9
 
 | | Crystal structure of fluorophore-labeled beta-lactamase PenP in complex with cefotaxime | | Descriptor: | 1-[6-(dimethylamino)naphthalen-2-yl]ethanone, Beta-lactamase, CEFOTAXIME, ... | | Authors: | Wong, W.-T, Zhao, Y.-X, Leung, Y.-C. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increased structural flexibility at the active site of a fluorophore-conjugated beta-lactamase distinctively impacts its binding toward diverse cephalosporin antibiotics
J.Biol.Chem., 286, 2011
|
|
3TG9
 
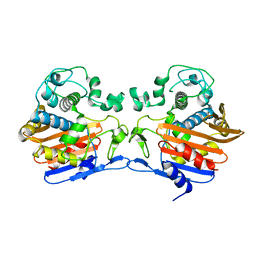 | | The crystal structure of penicillin binding protein from Bacillus halodurans | | Descriptor: | Penicillin-binding protein | | Authors: | Zhang, Z, Satyanarayana, L, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of penicillin binding protein from Bacillus halodurans
To be Published
|
|
3S1Y
 
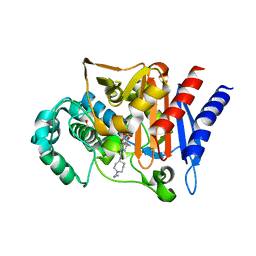 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with a beta-lactamase inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3S4X
 
 | |
3VWR
 
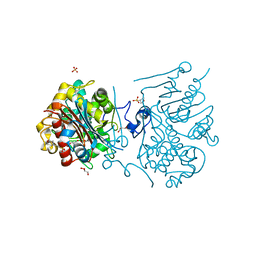 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187G/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWM
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187A/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWQ
 
 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
4N92
 
 | | Crystal structure of beta-lactamse PenP_E166S | | Descriptor: | Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4OKP
 
 | |
4N1H
 
 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
3VWP
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWN
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
4N9L
 
 | | crystal structure of beta-lactamse PenP_E166S in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
3VWL
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187S/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
