8P31
 
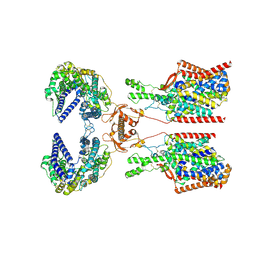 | | Structure of human SIT1:ACE2 complex (closed PD conformation) bound to L-pipecolate | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2X
 
 | | Structure of human SIT1:ACE2 complex (open PD conformation) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
1GNE
 
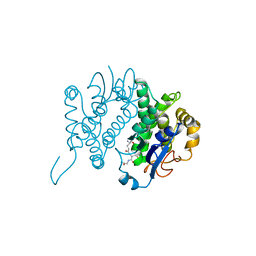 | | THE THREE-DIMENSIONAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE OF SCHISTOSOMA JAPONICUM FUSED WITH A CONSERVED NEUTRALIZING EPITOPE ON GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Lim, K, Ho, J.X, Keeling, K, Gilliland, G.L, Ji, X, Ruker, F, Carter, D.C. | | Deposit date: | 1994-06-16 | | Release date: | 1994-11-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV.
Protein Sci., 3, 1994
|
|
1G6V
 
 | | Complex of the camelid heavy-chain antibody fragment CAB-CA05 with bovine carbonic anhydrase | | Descriptor: | ANTIBODY HEAVY CHAIN, CARBONIC ANHYDRASE, ZINC ION | | Authors: | Desmyter, A, Decanniere, K, Muyldermans, S, Wyns, L. | | Deposit date: | 2000-11-08 | | Release date: | 2000-11-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody.
J.Biol.Chem., 276, 2001
|
|
1GEQ
 
 | |
1SZO
 
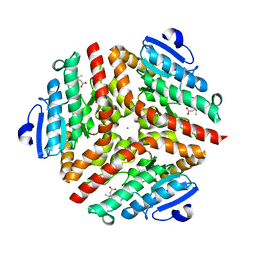 | | Crystal Structure Analysis of the 6-Oxo Camphor Hydrolase His122Ala Mutant Bound to Its Natural Product (2S,4S)-alpha-Campholinic Acid | | Descriptor: | (2S,4S)-4-(2,2-DIHYDROXYETHYL)-2,3,3-TRIMETHYLCYCLOPENTANONE, 6-oxocamphor hydrolase, CALCIUM ION | | Authors: | Leonard, P.M, Grogan, G. | | Deposit date: | 2004-04-06 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of 6-oxo camphor hydrolase H122A mutant bound to its natural product, (2S,4S)-alpha-campholinic acid: mutant structure suggests an atypical mode of transition state binding for a crotonase homolog.
J.Biol.Chem., 279, 2004
|
|
1A22
 
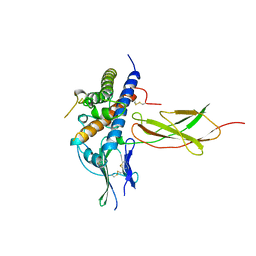 | | HUMAN GROWTH HORMONE BOUND TO SINGLE RECEPTOR | | Descriptor: | GROWTH HORMONE, GROWTH HORMONE RECEPTOR | | Authors: | De Vos, A.M, Ultsch, M. | | Deposit date: | 1998-01-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity.
J.Mol.Biol., 277, 1998
|
|
1PVJ
 
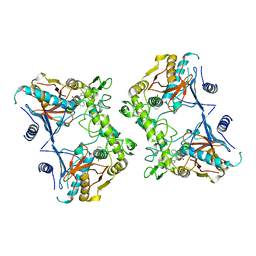 | | Crystal structure of the Streptococcal pyrogenic exotoxin B (SpeB)- inhibitor complex | | Descriptor: | (3R)-3-{[(BENZYLOXY)CARBONYL]AMINO}-2-OXO-4-PHENYLBUTANE-1-DIAZONIUM, pyrogenic exotoxin B | | Authors: | Ziomek, E, Sivaraman, J, Doran, J, Menard, R, Cygler, M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-09-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of autoprocessing of the streptococcal pyrogenic exotoxin B (speB). Crystal structure of the proenzyme-inhibitor complex
To be published
|
|
4REX
 
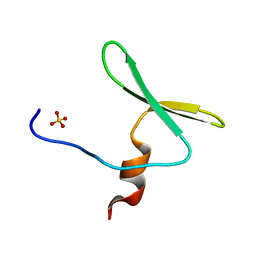 | |
1R6U
 
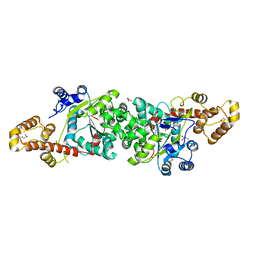 | | Crystal structure of an active fragment of human tryptophanyl-tRNA synthetase with cytokine activity | | Descriptor: | GLYCEROL, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Yang, X.-L, Otero, F.J, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-10-16 | | Release date: | 2004-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase
Structure, 15, 2007
|
|
3R0L
 
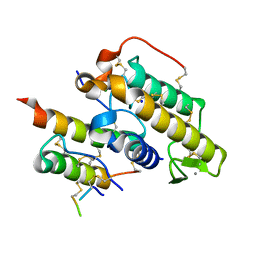 | | Crystal structure of crotoxin | | Descriptor: | ACETATE ION, CHLORIDE ION, Crotoxin chain A, ... | | Authors: | Saul, F.A, Faure, G. | | Deposit date: | 2011-03-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Crotoxin Reveals Key Residues Involved in the Stability and Toxicity of This Potent Heterodimeric Beta-Neurotoxin
J.Mol.Biol., 412, 2011
|
|
1SWH
 
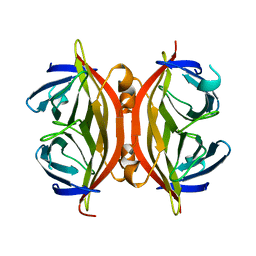 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWN
 
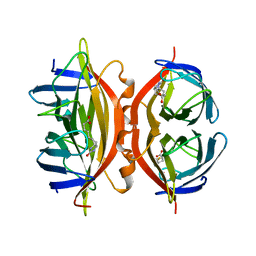 | | CORE-STREPTAVIDIN MUTANT W108F IN COMPLEX WITH BIOTIN AT PH 7.0 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWQ
 
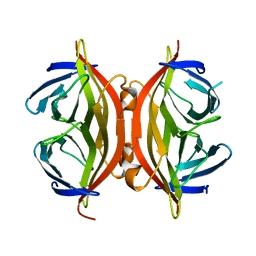 | | CORE-STREPTAVIDIN MUTANT W120A AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWP
 
 | | CORE-STREPTAVIDIN MUTANT W120F IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWJ
 
 | | CORE-STREPTAVIDIN MUTANT W79F AT PH 4.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWO
 
 | | CORE-STREPTAVIDIN MUTANT W120F AT PH 7.5 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWK
 
 | | CORE-STREPTAVIDIN MUTANT W79F IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWL
 
 | | CORE-STREPTAVIDIN MUTANT W108F AT PH 7.0 | | Descriptor: | CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
1SWR
 
 | | CORE-STREPTAVIDIN MUTANT W120A IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
2YGP
 
 | | WIF domain-EGF-like domain 1 Met77Trp of human Wnt inhibitory factor 1 in complex with 1,2-dipalmitoylphosphatidylcholine | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Malinauskas, T, Aricescu, A.R, Lu, W, Siebold, C, Jones, E.Y. | | Deposit date: | 2011-04-19 | | Release date: | 2011-07-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Modular Mechanism of Wnt Signaling Inhibition by Wnt Inhibitory Factor 1
Nat.Struct.Mol.Biol., 18, 2011
|
|
1AEV
 
 | | INTRODUCTION OF NOVEL SUBSTRATE OXIDATION INTO CYTOCHROME C PEROXIDASE BY CAVITY COMPLEMENTATION: OXIDATION OF 2-AMINOTHIAZOLE AND COVALENT MODIFICATION OF THE ENZYME (2-AMINOTHIAZOLE) | | Descriptor: | 2-AMINOTHIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Fitzgerald, M.M, Jensen, G.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Introduction of novel substrate oxidation into cytochrome c peroxidase by cavity complementation: oxidation of 2-aminothiazole and covalent modification of the enzyme.
Biochemistry, 36, 1997
|
|
8DOV
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 (HID2) in complex with Hemoglobin | | Descriptor: | GLYCEROL, Heme-binding protein Shr, Hemoglobin subunit alpha, ... | | Authors: | Macdonald, R, Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1ALZ
 
 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | ETHANOL, ILE-GRAMICIDIN C, VAL-GRAMICIDIN A | | Authors: | Burkhart, B.M, Pangborn, W.A, Duax, W.L, Langs, D.A. | | Deposit date: | 1997-06-06 | | Release date: | 1998-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
