3V72
 
 | | Crystal Structure of Rat DNA polymerase beta Mutator E295K: Enzyme-dsDNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(P*AP*AP*AP*CP*TP*CP*AP*CP*AP*T)-3', DNA 5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3', ... | | Authors: | Gridley, C.L, Jaeger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Unfavorable Electrostatic and Steric Interactions in DNA Polymerase beta E295K Mutant Interfere with the Enzyme s Pathway
J.Am.Chem.Soc., 134, 2012
|
|
2PQ8
 
 | | MYST histone acetyltransferase 1 | | Descriptor: | COENZYME A, Probable histone acetyltransferase MYST1, UNKNOWN ATOM OR ION, ... | | Authors: | Tempel, W, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MYST histone acetyltransferase 1.
To be Published
|
|
5AZA
 
 | |
7KTX
 
 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to a Fab in DDM detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
5FFT
 
 | | Crystal Structure of Surfactant Protein-A Y221A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
8K3Z
 
 | | Cryo-EM structure of CXCR4 in complex with CXCL12 | | Descriptor: | C-X-C chemokine receptor type 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Y.Z, Liu, A.J, Liao, Q.W, Ye, R.D. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of CXCR4 in complex with CXCL12
To Be Published
|
|
5HV8
 
 | | Solution structure of an octanoyl- loaded acyl carrier protein domain from module MLSA2 of the mycolactone polyketide synthase. | | Descriptor: | S-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] octanethioate, Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
8K4P
 
 | |
7KSR
 
 | | PRC2:EZH1_A from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KSO
 
 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KZO
 
 | | Outer dynein arm docking complex bound to doublet microtubules from C. reinhardtii | | Descriptor: | Dynein gamma chain, flagellar outer arm, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
7KZM
 
 | | Outer dynein arm bound to doublet microtubules from C. reinhardtii | | Descriptor: | DC1, DC2, Dynein 11 kDa light chain, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
2OZM
 
 | | Crystal structure of RB69 gp43 in complex with DNA with 5-NITP opposite an abasic site analog | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO -1H-INDOLE, DNA polymerase, MAGNESIUM ION, ... | | Authors: | Zahn, K.E, Belrhali, H, Wallace, S.S, Doublie, S. | | Deposit date: | 2007-02-26 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Caught Bending the A-Rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide
Biochemistry, 46, 2007
|
|
7KVA
 
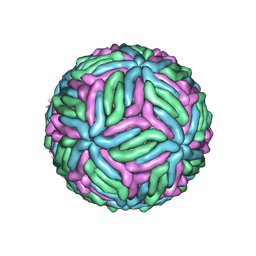 | | Structure of West Nile virus (Kunjin) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Matrix protein M | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-11-27 | | Release date: | 2020-12-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A unified route for flavivirus structures uncovers essential pocket factors conserved across pathogenic viruses.
Nat Commun, 12, 2021
|
|
7MEZ
 
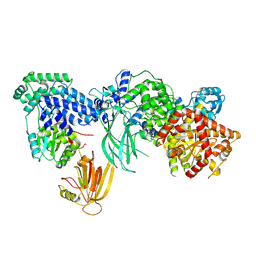 | | Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 5 | | Authors: | Burke, J.E, Dalwadi, U, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the phosphoinositide 3-kinase (PI3K) p110 gamma-p101 complex reveals molecular mechanism of GPCR activation.
Sci Adv, 7, 2021
|
|
6IAC
 
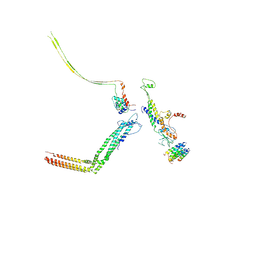 | | Portal and tail of native bacteriophage P68 | | Descriptor: | Lower collar protein, Minor structural protein, Portal protein, ... | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-26 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6IAW
 
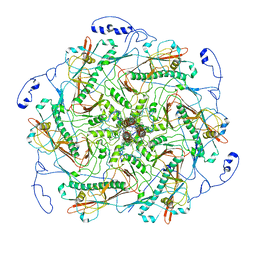 | | Structure of head fiber and inner core protein gp22 of native bacteriophage P68 | | Descriptor: | Arstotzka protein, Head fiber, Major head protein, ... | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
5H64
 
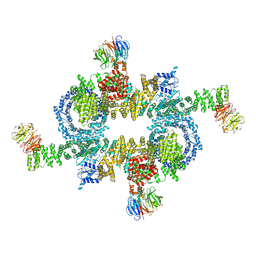 | | Cryo-EM structure of mTORC1 | | Descriptor: | Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Yang, H, Wang, J, Liu, M, Chen, X, Huang, M, Tan, D, Dong, M, Wong, C.C.L, Wang, J, Xu, Y, Wang, H. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-25 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | 4.4 angstrom Resolution Cryo-EM structure of human mTOR Complex 1
Protein Cell, 7, 2016
|
|
2OZS
 
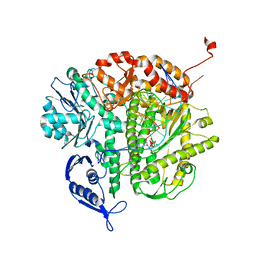 | | Crystal structure of RB69 gp43 in complex with DNA with dATP opposite dTMP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, MAGNESIUM ION, ... | | Authors: | Zahn, K.E, Belrhali, H, Wallace, S.S, Doublie, S. | | Deposit date: | 2007-02-27 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Caught bending the a-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide.
Biochemistry, 46, 2007
|
|
5FEK
 
 | |
7M30
 
 | |
7M22
 
 | |
7KZN
 
 | | Outer dynein arm core subcomplex from C. reinhardtii | | Descriptor: | DC1, DC2, Dynein 11 kDa light chain, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
8BMW
 
 | | SsoCsm | | Descriptor: | CRISPR-associated Cas7 paralog (Type III-D), CRISPR-associated protein Cas10 (Type III-D), CRISPR-associated protein Cas5 (Type III-D), ... | | Authors: | Spagnolo, L, White, M.F. | | Deposit date: | 2022-11-11 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Saccharolobus solfataricus type III-D CRISPR effector.
Curr Res Struct Biol, 5, 2023
|
|
7KR5
 
 | | Cryo-EM structure of the CRAC channel Orai in an open conformation; H206A gain-of-function mutation in complex with an antibody | | Descriptor: | 19B5 Fab heavy chain, 19B5 Fab light chain, CALCIUM ION, ... | | Authors: | Long, S.B, Hou, X, Outhwaite, I.R. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the calcium release-activated calcium channel Orai in an open conformation.
Elife, 9, 2020
|
|
