5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
1A38
 
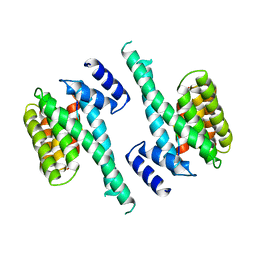 | | 14-3-3 PROTEIN ZETA BOUND TO R18 PEPTIDE | | Descriptor: | 14-3-3 PROTEIN ZETA, R18 PEPTIDE (PHCVPRDLSWLDLEANMCLP) | | Authors: | Petosa, C, Masters, S.C, Pohl, J, Wang, B, Fu, H, Liddington, R.C. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove.
J.Biol.Chem., 273, 1998
|
|
3PVD
 
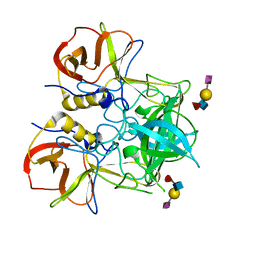 | | Crystal structure of P domain dimer of Norovirus VA207 complexed with 3'-sialyl-Lewis x tetrasaccharide | | Descriptor: | Capsid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
4RY3
 
 | |
4HFD
 
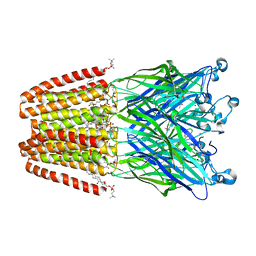 | | The GLIC pentameric Ligand-Gated Ion Channel F14'A ethanol-sensitive mutant complexed to bromoform | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Howard, R.J, Malherbe, L, Lee, U.S, Corringer, P.J, Harris, R.A, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel.
Nat Commun, 4, 2013
|
|
6CIP
 
 | |
5LU1
 
 | | Human 14-3-3 sigma CLU3 mutant complexed with short HSPB6 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heat shock protein beta-6 | | Authors: | Sluchanko, N.N, Beelen, S, Kulikova, A.A, Weeks, S.D, Antson, A.A, Gusev, N.B, Strelkov, S.V. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Structure, 25, 2017
|
|
3PVM
 
 | | Structure of Complement C5 in Complex with CVF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, Complement C5 | | Authors: | Laursen, N.S, Andersen, K.R, Braren, I, Sottrup-Jensen, L, Spillner, E, Andersen, G.R. | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex.
Embo J., 30, 2011
|
|
4RYZ
 
 | |
8ENR
 
 | |
8OUG
 
 | |
5LY7
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Beta-hexosaminidase, DI(HYDROXYETHYL)ETHER | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-09-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
1ADX
 
 | | FIFTH EGF-LIKE DOMAIN OF THROMBOMODULIN (TMEGF5), NMR, 14 STRUCTURES | | Descriptor: | THROMBOMODULIN | | Authors: | Sampoli-Benitez, B.A, Hunter, M.J, Meininger, D.P, Komives, E.A. | | Deposit date: | 1997-02-18 | | Release date: | 1997-12-24 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the fifth EGF-like domain of thrombomodulin: An EGF-like domain with a novel disulfide-bonding pattern.
J.Mol.Biol., 273, 1997
|
|
1KJ1
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM GARLIC (ALLIUM SATIVUM) BULBS COMPLEXED WITH ALPHA-D-MANNOSE | | Descriptor: | alpha-D-mannopyranose, lectin I, lectin II | | Authors: | Ramachandraiah, G, Chandra, N.R, Surolia, A, Vijayan, M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-02-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Re-refinement using reprocessed data to improve the quality of the structure: a case study involving garlic lectin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4S2X
 
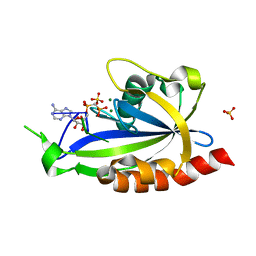 | | Structure of E. coli RppH bound to RNA and two magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), RNA pyrophosphohydrolase, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
5LZ9
 
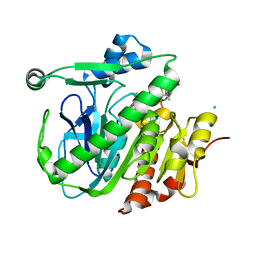 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 2-fluoranyl-5-[2-[(4~{S})-4-[4-methyl-1,1-bis(oxidanylidene)thian-4-yl]-2-oxidanylidene-pyrrolidin-1-yl]ethoxy]benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase, ... | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
4H3X
 
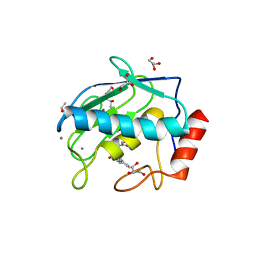 | | Crystal structure of an MMP broad spectrum hydroxamate based inhibitor CC27 in complex with the MMP-9 catalytic domain | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
8CKS
 
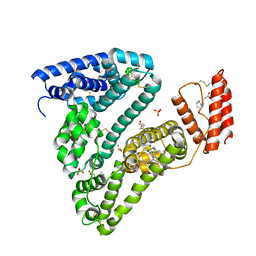 | | Crystal structure of Human Serum Albumin in complex with FESAN | | Descriptor: | 3,3'-commo-bis(1,2-dicarba-3-ferra-closo-dodecaborane), DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Dolot, R.M, Kaniowski, D, Ebenryter-Olbinska, K, Szczupak, P, Suwara, J, Nawrot, B.C. | | Deposit date: | 2023-02-16 | | Release date: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Serum Albumin in complex with FESAN
To Be Published
|
|
1AN9
 
 | | D-AMINO ACID OXIDASE COMPLEX WITH O-AMINOBENZOATE | | Descriptor: | 2-AMINOBENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Miura, R, Setoyama, C, Nishina, Y, Shiga, K, Mizutani, H, Miyahara, I, Hirotsu, K. | | Deposit date: | 1997-06-28 | | Release date: | 1997-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic studies on D-amino acid oxidase x substrate complex: implications of the crystal structure of enzyme x substrate analog complex.
J.Biochem.(Tokyo), 122, 1997
|
|
4HD8
 
 | | Crystal structure of human Sirt3 in complex with Fluor-de-Lys peptide and piceatannol | | Descriptor: | Fluor-de-Lys peptide, ISOPROPYL ALCOHOL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Steegborn, C. | | Deposit date: | 2012-10-02 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A molecular mechanism for direct sirtuin activation by resveratrol.
Plos One, 7, 2012
|
|
1T6P
 
 | |
5T31
 
 | | Exploiting an Asp-Glu switch in Glycogen Synthase Kinase 3 to design paralog selective inhibitors for use in acute myeloid leukemia | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Stein, A.J, Holson, E.B, Wagner, F.F, Cambell, A.J. | | Deposit date: | 2016-08-24 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
4RRW
 
 | | Crystal Structure of Apo Murine Cyclooxygenase-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prostaglandin G/H synthase 2, ... | | Authors: | Xu, S, Blobaum, A.L, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Action at a Distance: MUTATIONS OF PERIPHERAL RESIDUES TRANSFORM RAPID REVERSIBLE INHIBITORS TO SLOW, TIGHT BINDERS OF CYCLOOXYGENASE-2.
J.Biol.Chem., 290, 2015
|
|
4TMV
 
 | |
1AIK
 
 | | HIV GP41 CORE STRUCTURE | | Descriptor: | HIV-1 GP41 GLYCOPROTEIN | | Authors: | Chan, D.C, Fass, D, Berger, J.M, Kim, P.S. | | Deposit date: | 1997-04-20 | | Release date: | 1997-06-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Core structure of gp41 from the HIV envelope glycoprotein.
Cell(Cambridge,Mass.), 89, 1997
|
|
