7E33
 
 | | Serotonin 1E (5-HT1E) receptor-Gi protein complex | | Descriptor: | 3-(1-methylpiperidin-4-yl)-1H-indol-5-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Y
 
 | | Serotonin-bound Serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E2Z
 
 | | Aripiprazole-bound serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 7-[4-[4-[2,3-bis(chloranyl)phenyl]piperazin-1-yl]butoxy]-3,4-dihydro-1H-quinolin-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E32
 
 | | Serotonin 1D (5-HT1D) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
7E6H
 
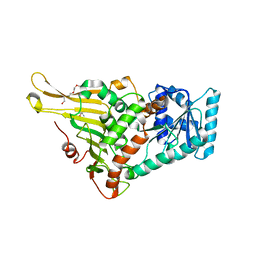 | | glucose-6-phosphate dehydrogenase from Kluyveromyces lactis | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Ha, V.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
7E6I
 
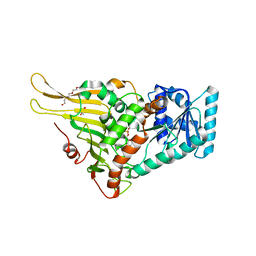 | | Glucose-6-phosphate dehydrogenase in complex with its substrate glucose-6-phosphate | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Vu, H.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
5KP4
 
 | | Crystal Structure of Ketosteroid Isomerase from Pseudomonas putida (pKSI) bound to 19-nortestosterone | | Descriptor: | (8~{R},9~{S},10~{R},13~{S},14~{S},17~{S})-13-methyl-17-oxidanyl-2,6,7,8,9,10,11,12,14,15,16,17-dodecahydro-1~{H}-cyclop enta[a]phenanthren-3-one, Steroid Delta-isomerase | | Authors: | Wu, Y, Boxer, S.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.706 Å) | | Cite: | A Critical Test of the Electrostatic Contribution to Catalysis with Noncanonical Amino Acids in Ketosteroid Isomerase.
J.Am.Chem.Soc., 138, 2016
|
|
7E2X
 
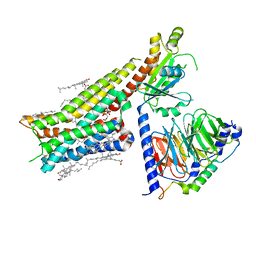 | | Apo serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
6N94
 
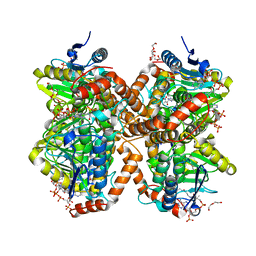 | | Methylmalonyl-CoA decarboxylase in complex with 2-nitronate-propionyl-amino(dethia)-CoA | | Descriptor: | DI(HYDROXYETHYL)ETHER, IMIDAZOLE, Methylmalonyl-CoA decarboxylase, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
7E80
 
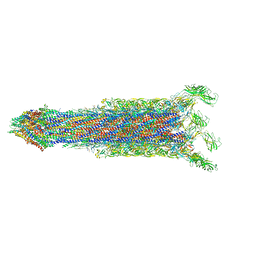 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E90
 
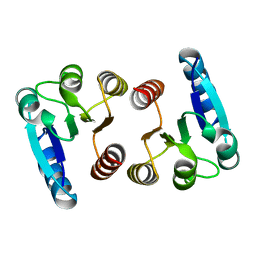 | |
7E92
 
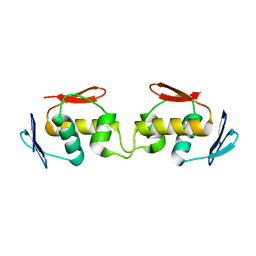 | |
7E82
 
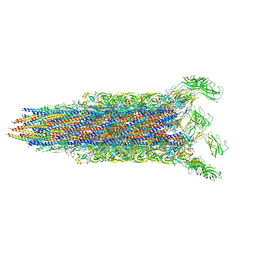 | | Cryo-EM structure of the flagellar rod with partial hook from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
2X7C
 
 | | Crystal structure of human kinesin Eg5 in complex with (S)-enastron | | Descriptor: | (S)-4-(3-HYDROXYPHENYL)-2-THIOXO-1,2,3,4,7,8-HEXAHYDROQUINAZOLIN-5(6H)-ONE, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Kaan, H.Y.K, Ulaganathan, V, Rath, O, Laggner, C, Prokopcova, H, Dallinger, D, Kappe, C.O, Kozielski, F. | | Deposit date: | 2010-02-26 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhibition of Eg5 by Dihydropyrimidines: Stereoselectivity of Antimitotic Inhibitors Enastron, Dimethylenastron and Fluorastrol.
J.Med.Chem., 53, 2010
|
|
7PWI
 
 | | Structure of the dTDP-sugar epimerase StrM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, dTDP-4-keto-rhamnose 3,5-epimerase,dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Cross, A.R, Harmer, N.J, Isupov, M.N. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.326 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases.
J.Biol.Chem., 298, 2022
|
|
7ELC
 
 | | Structure of monomeric complex of MACV L bound to Z and 3'-vRNA | | Descriptor: | 3'-vRNA promoter, MANGANESE (II) ION, RING finger protein Z, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
7ELA
 
 | | Structure of Lassa virus polymerase in complex with 3'-vRNA and Z mutant (F36A) | | Descriptor: | 3-'vRNA promoter, MANGANESE (II) ION, RING finger protein Z, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
3PK1
 
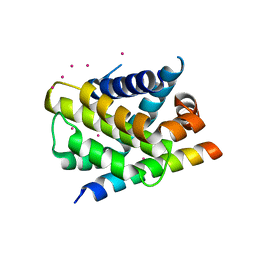 | | Crystal structure of Mcl-1 in complex with the BaxBH3 domain | | Descriptor: | Apoptosis regulator BAX, CADMIUM ION, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2010-11-11 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Mutation to Bax beyond the BH3 domain disrupts interactions with pro-survival proteins and promotes apoptosis
J.Biol.Chem., 286, 2011
|
|
2XAC
 
 | |
7DWM
 
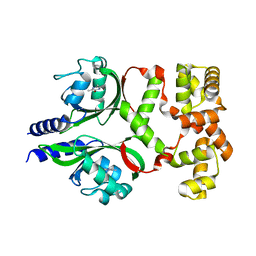 | | Crystal structure of the phage VqmA-DPO complex | | Descriptor: | 3,5-dimethylpyrazin-2-ol, Transcriptional regulator | | Authors: | Gu, Y, Yang, W.S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Understanding the mechanism of asymmetric gene regulation determined by the VqmA of vibriophage.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
4N9M
 
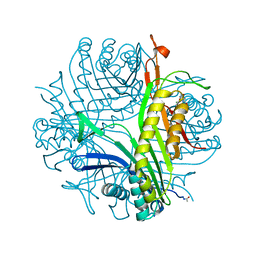 | | Joint neutron/x-ray structure of urate oxidase in complex with 8-hydroxyxanthine | | Descriptor: | 8-hydroxy-3,9-dihydro-1H-purine-2,6-dione, CHLORIDE ION, SODIUM ION, ... | | Authors: | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | NEUTRON DIFFRACTION (2.023 Å), X-RAY DIFFRACTION | | Cite: | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
7EB2
 
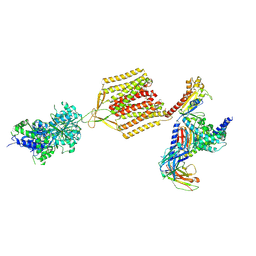 | | Cryo-EM structure of human GABA(B) receptor-Gi protein complex | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Shen, C, Mao, C, Xu, C, Jin, N, Zhang, H, Shen, D, Shen, Q, Wang, X, Hou, T, Rondard, P, Chen, Z, Pin, J, Zhang, Y, Liu, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of GABA B receptor-G i protein coupling.
Nature, 594, 2021
|
|
7SA5
 
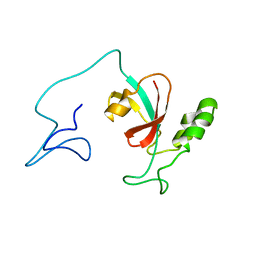 | | Two-state solution NMR structure of Apo Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Born, A, Vogeli, B. | | Deposit date: | 2021-09-22 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Reconstruction of Coupled Intra- and Interdomain Protein Motion from Nuclear and Electron Magnetic Resonance.
J.Am.Chem.Soc., 143, 2021
|
|
2WYJ
 
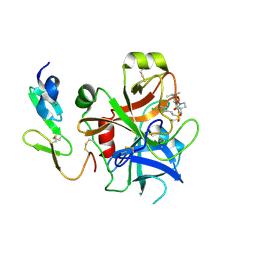 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1S)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
