7V3U
 
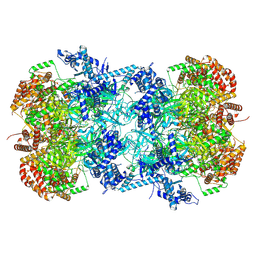 | | Cryo-EM structure of MCM double hexamer with structured Mcm4-NSD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7SQL
 
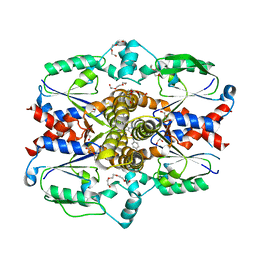 | | Crystal structure of human uridine-cytidine kinase 2 complexed with a weak small molecule inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-(4-bromophenyl)-2-{[1-(4-fluorophenyl)-4-oxo-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-6-yl]sulfanyl}acetamide, ... | | Authors: | Mashayekh, S, Stunkard, L.M, Kienle, M, Mathews, I.I, Khosla, C. | | Deposit date: | 2021-11-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prototyping of Allosteric Inhibitors of Human Uridine/Cytidine Kinase 2 (UCK2).
Biochemistry, 61, 2022
|
|
7E2M
 
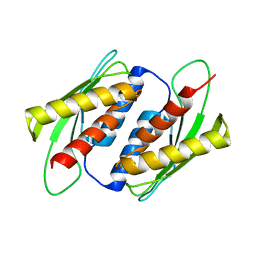 | | Crystal structure of the RWD domain of human GCN2 - 2 | | Descriptor: | eIF-2-alpha kinase GCN2 | | Authors: | Hei, Z, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures reveal a novel dimer of the RWD domain of human general control nonderepressible 2.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
5LI7
 
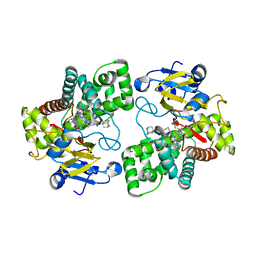 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with 1-(3-(1H-imidazol-1-yl)propyl)-3-((3s,5s,7s)-adamantan-1-yl)urea | | Descriptor: | 1-(1-adamantyl)-3-(3-imidazol-1-ylpropyl)urea, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126 | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
7T8B
 
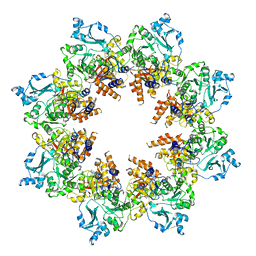 | | Octameric Human Twinkle Helicase Clinical Variant W315L | | Descriptor: | Twinkle mtDNA helicase | | Authors: | Riccio, A.A, Bouvette, J, Krahn, J, Borgnia, M.J, Copeland, W.C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight and characterization of human Twinkle helicase in mitochondrial disease.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SUS
 
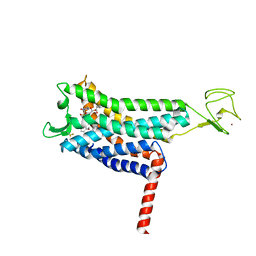 | | Crystal structure of Apelin receptor in complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Han, G.W, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7E2K
 
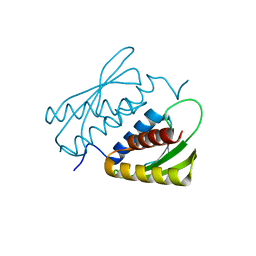 | | Crystal structure of the RWD domain of human GCN2 - 1 | | Descriptor: | eIF-2-alpha kinase GCN2 | | Authors: | Hei, Z, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Crystal structures reveal a novel dimer of the RWD domain of human general control nonderepressible 2.
Biochem.Biophys.Res.Commun., 549, 2021
|
|
7DTK
 
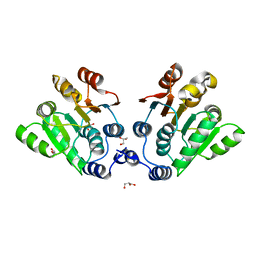 | |
7DTJ
 
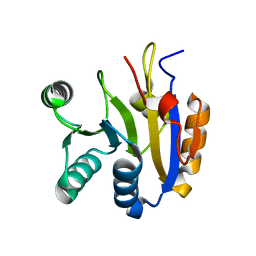 | |
7V3V
 
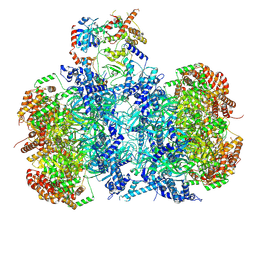 | | Cryo-EM structure of MCM double hexamer bound with DDK in State I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 7, DDK kinase regulatory subunit DBF4, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
7SN8
 
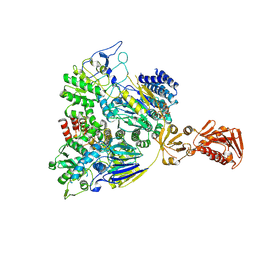 | |
5V68
 
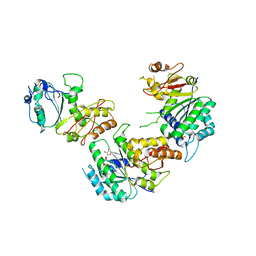 | | Crystal structure of cell division protein FtsZ from Mycobacterium tuberculosis bounded via the T9 loop | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Lazo, E.O, Ojima, I, Chowdhury, S.R, Awasthi, D, Jakoncic, J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Novel T9 loop conformation of filamenting temperature-sensitive mutant Z from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7BZW
 
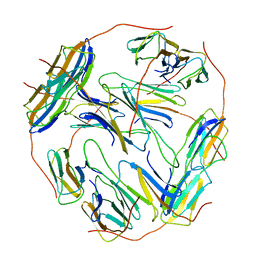 | | Structure of Hsp21 | | Descriptor: | Heat shock protein 21, chloroplastic | | Authors: | Lau, W.C.Y. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of substrate recognition and thermal protection by a small heat shock protein.
Nat Commun, 12, 2021
|
|
5LHF
 
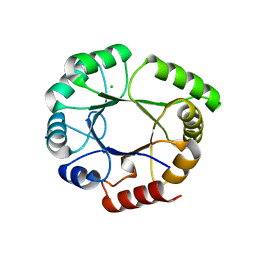 | |
7ZSC
 
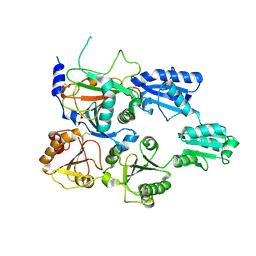 | | Crystal structure of the heterodimeric human C-P4H-II with truncated alpha subunit (C-P4H-II delta281) | | Descriptor: | Prolyl 4-hydroxylase subunit alpha-2, Protein disulfide-isomerase, SULFATE ION | | Authors: | Lebedev, A, Koski, M.K, Wierenga, R.K, Murthy, A.V, Sulu, R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the collagen prolyl 4-hydroxylase (C-P4H) catalytic domain complexed with PDI: Toward a model of the C-P4H alpha 2 beta 2 tetramer.
J.Biol.Chem., 298, 2022
|
|
5UQK
 
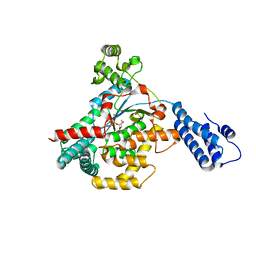 | |
4ZHU
 
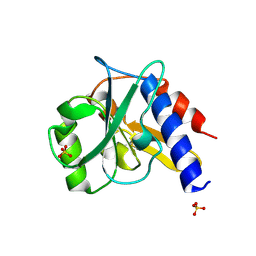 | | Crystal structure of a bacterial repressor protein | | Descriptor: | SULFATE ION, YfiR | | Authors: | Li, S, Li, T, Wang, Y, Bartlam, M. | | Deposit date: | 2015-04-27 | | Release date: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.3968 Å) | | Cite: | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
7ZQ9
 
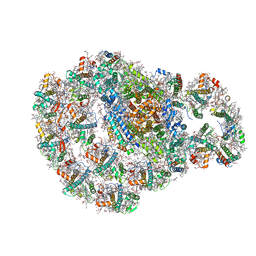 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7T6X
 
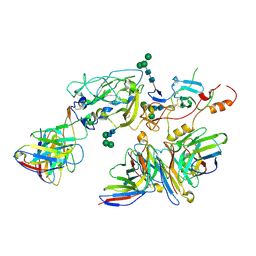 | | Cryo-EM structure of full-length hepatitis C virus E1E2 glycoprotein in complex with AR4A, AT12009, and IGH505 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AR4A Fab, ... | | Authors: | Torrents de la Pena, A, Ward, A.B, Eshun-Wilson, L, Lander, G.C. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the hepatitis C virus E1E2 glycoprotein complex.
Science, 378, 2022
|
|
5LFJ
 
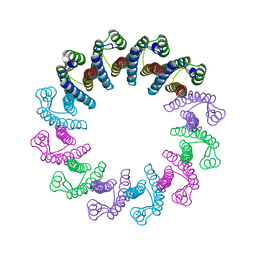 | | Crystal Structure of the Bacterial Proteasome Activator Bpa of Mycobacterium tuberculosis | | Descriptor: | Bacterial proteasome activator | | Authors: | Bolten, M, Delley, C.L, Leibundgut, M, Boehringer, D, Ban, N, Weber-Ban, E. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Bacterial Proteasome Activator Bpa in Complex with the 20S Proteasome.
Structure, 24, 2016
|
|
5UK6
 
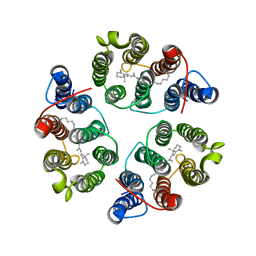 | | Structure of Anabaena Sensory Rhodopsin Determined by Solid State NMR Spectroscopy and DEER | | Descriptor: | Bacteriorhodopsin | | Authors: | Milikisiyants, S, Wang, S, Munro, R.A, Donohue, M, Ward, M.E, Brown, L.S, Smirnova, T.I, Ladizhansky, V, Smirnov, A.I. | | Deposit date: | 2017-01-20 | | Release date: | 2017-05-31 | | Last modified: | 2020-01-08 | | Method: | SOLID-STATE NMR | | Cite: | Oligomeric Structure of Anabaena Sensory Rhodopsin in a Lipid Bilayer Environment by Combining Solid-State NMR and Long-range DEER Constraints.
J. Mol. Biol., 429, 2017
|
|
7FFP
 
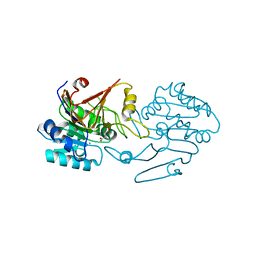 | | Crystal structure of di-peptidase-E from Xenopus laevis | | Descriptor: | ASPARTIC ACID, Alpha-aspartyl dipeptidase, CALCIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
6ZRC
 
 | |
7FFT
 
 | |
7FFW
 
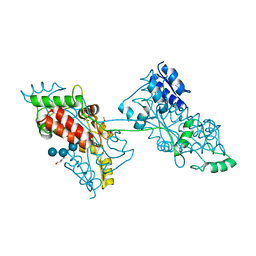 | | The crystal structure of a domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, METHOXYETHANE, ... | | Authors: | Wang, L, Bu, T, Bai, X. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
