4WJZ
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG)(G141A) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, PHOSPHATE ION | | Authors: | Hou, J, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
3JS5
 
 | |
3JSJ
 
 | |
4WHE
 
 | | Crystal structure of E. coli phage shock protein A (PspA 1-144) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phage shock protein A | | Authors: | Parthier, C, Schoepfel, M, Stubbs, M.T, Osadnik, H, Brueser, T. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PspF-binding domain PspA1-144 and the PspAF complex: New insights into the coiled-coil-dependent regulation of AAA+ proteins.
Mol.Microbiol., 98, 2015
|
|
4WHJ
 
 | |
3K93
 
 | |
4WFA
 
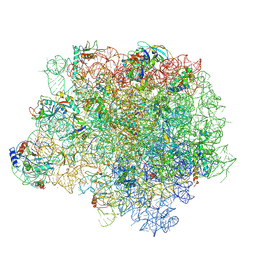 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with linezolid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.392 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WHX
 
 | | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Branched-chain-amino-acid transaminase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fairman, J.W, Dranow, D.M, Taylor, B.M, Lorimer, D, Edwards, T.E. | | Deposit date: | 2014-09-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystal Structure of an Amino Acid Aminotransferase from Burkholderia pseudomallei Bound to the Co-factor Pyridoxal Phosphate
to be published
|
|
3K9J
 
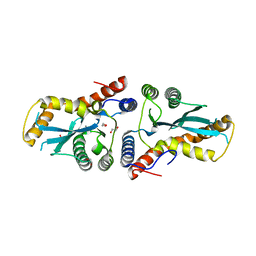 | | Transposase domain of Metnase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Histone-lysine N-methyltransferase SETMAR | | Authors: | Goodwin, K.D, He, H, Imasaki, T, Lee, S.-H, Georgiadis, M.M. | | Deposit date: | 2009-10-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of the human Hsmar1-derived transposase domain in the DNA repair enzyme Metnase.
Biochemistry, 49, 2010
|
|
3JV4
 
 | |
4WKB
 
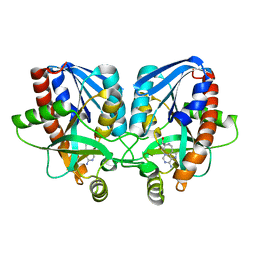 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2014-10-02 | | Release date: | 2015-08-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
4WJ5
 
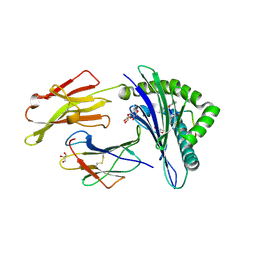 | | Structure of HLA-A2 in complex with an altered peptide ligands based on Mart-1 variant epitope | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Celie, P.H.N, Rodenko, B, Ovaa, H. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Altered Peptide Ligands Revisited: Vaccine Design through Chemically Modified HLA-A2-Restricted T Cell Epitopes.
J Immunol., 193, 2014
|
|
3KA9
 
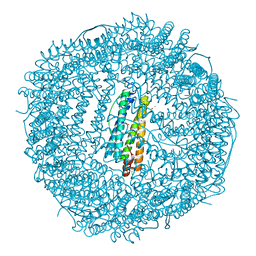 | | Frog M-ferritin, EEH mutant, with cobalt | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferritin, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-19 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Frog M-ferritin, EEH mutant, with cobalt
To be Published
|
|
4WJM
 
 | |
4WOQ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric | | Descriptor: | 2-KETOBUTYRIC ACID, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-16 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric
to be published
|
|
3S11
 
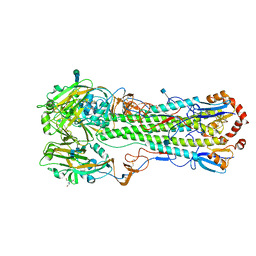 | | Crystal structure of H5N1 influenza virus hemagglutinin, strain 437-10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | DuBois, R.M, Zaraket, H, Reddivari, M, Heath, R.J, White, S.W, Russell, C.J. | | Deposit date: | 2011-05-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity.
Plos Pathog., 7, 2011
|
|
4WK5
 
 | | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458 | | Descriptor: | Geranyltranstransferase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a Isoprenoid Synthase family member from Thermotoga neapolitana DSM 4359, target EFI-509458
To be published
|
|
3JYS
 
 | |
3K01
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
3K1N
 
 | |
3K22
 
 | | Glucocorticoid Receptor with Bound alaninamide 10 with TIF2 peptide | | Descriptor: | Glucocorticoid receptor, N-[(1R)-2-amino-1-methyl-2-oxoethyl]-3-(6-methyl-4-{[3,3,3-trifluoro-2-hydroxy-2-(trifluoromethyl)propyl]amino}-1H-indazol-1-yl)benzamide, Transcriptional Intermediary Factor 2, ... | | Authors: | Biggadike, K.B, McLay, I.M, Madauss, K.P, Williams, S.P, Bledsoe, R.K. | | Deposit date: | 2009-09-29 | | Release date: | 2010-08-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K3Q
 
 | | Crystal Structure of a Llama Antibody complexed with the C. Botulinum Neurotoxin Serotype A Catalytic Domain | | Descriptor: | Botulinum neurotoxin type A, ZINC ION, llama Aa1 VHH domain | | Authors: | Thompson, A.A, Dong, J, Marks, J.D, Stevens, R.C. | | Deposit date: | 2009-10-04 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Single-Domain Llama Antibody Potently Inhibits the Enzymatic Activity of Botulinum Neurotoxin by Binding to the Non-Catalytic alpha-Exosite Binding Region.
J.Mol.Biol., 397, 2010
|
|
3K46
 
 | |
3K4O
 
 | |
3S5Z
 
 | | Pharmacological Chaperoning in Human alpha-Galactosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, GLYCEROL, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2011-05-23 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The molecular basis of pharmacological chaperoning in human alpha-galactosidase
Chem.Biol., 18, 2011
|
|
