2XSR
 
 | | Crystal structure of wild type Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
7VKZ
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with 1-Deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, CALCIUM ION, beta-1,2-glucosyltransferase | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J.Biol.Chem., 298, 2022
|
|
5OU0
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 37 | | Descriptor: | 2-[5-(4-chlorophenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
3H01
 
 | |
5FI4
 
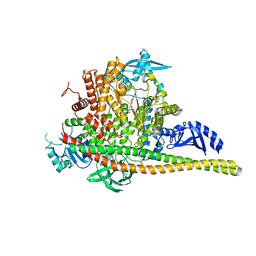 | | Discovery of imidazo[1,2-a]-pyridine inhibitors of pan-PI3 kinases that are efficacious in a mouse xenograft model | | Descriptor: | GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Elling, R.A, Knapp, M.S, Han, W, Daniel, L.M, Xy, Y, Burger, M.T, Ni, Z, Smith, A, Lan, J, Williams, T, Verhagen, J, Huh, K, Merritt, H, Chan, J, Kaufman, S, Voliva, C.F, Pecchi, S. | | Deposit date: | 2015-12-22 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of imidazo[1,2-a]-pyridine inhibitors of pan-PI3 kinases that are efficacious in a mouse xenograft model.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
4MBL
 
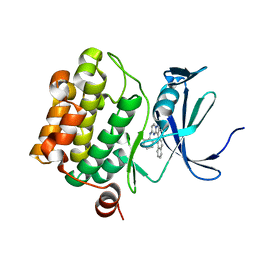 | | Discovery of Pyrazolo[1,5a]pyrimidine-based Pim1 Inhibitors | | Descriptor: | (1R,2R)-N-[3-(naphthalen-2-yl)pyrazolo[1,5-a]pyrimidin-5-yl]cyclohexane-1,2-diamine, Serine/threonine-protein kinase pim-1 | | Authors: | Azevedo, R, Fischmann, T.O. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of pyrazolo[1,5-a]pyrimidine-based Pim inhibitors: A template-based approach.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5T8H
 
 | |
2XSV
 
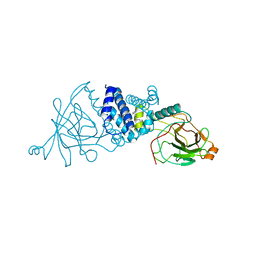 | | Crystal structure of L69A mutant Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
2Y1E
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese. | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, SULFATE ION | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
2XSU
 
 | | Crystal structure of the A72G mutant of Acinetobacter radioresistens catechol 1,2 dioxygenase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, CATECHOL 1,2 DIOXYGENASE, FE (III) ION | | Authors: | Micalella, C, Martignon, S, Bruno, S, Rizzi, M. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystallography, Mass Spectrometry and Single Crystal Microspectrophotometry: A Multidisciplinary Characterization of Catechol 1,2 Dioxygenase.
Biochim.Biophys.Acta, 1814, 2011
|
|
6BI9
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,2,5-trimethyl-1H-pyrrole-3-carboxylic acid | | Descriptor: | 1,2,5-trimethyl-1H-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,2,5-trimethyl-1H-pyrrole-3-carboxylic acid
To Be Published
|
|
2Y1C
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese. | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
3TH9
 
 | | Crystal Structure of HIV-1 Protease Mutant Q7K V32I L63I with a cyclic sulfonamide inhibitor | | Descriptor: | Gag-Pol polyprotein, tert-butyl {(2S,3R)-4-[(4S)-7-fluoro-4-methyl-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate | | Authors: | Orth, P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design, Synthesis, and X-ray Crystallographic Analysis of a Novel Class of HIV-1 Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
7FE3
 
 | |
6LNK
 
 | | Candida albicans Fructose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, Fructose-bisphosphate aldolase, ZINC ION | | Authors: | Huang, Y, Cao, H, Ren, Y, Wan, J. | | Deposit date: | 2019-12-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
3K2C
 
 | |
6B4N
 
 | | a hydroxymethyl functionality at the 4-position of the 2-phenyloxazole moiety of HIV-1 protease inhibitors involving the P2' ligands | | Descriptor: | CHLORIDE ION, Protease, SODIUM ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and X-ray Studies of HIV-1 Protease Inhibitors with Modified P2' Ligands of Darunavir.
ChemMedChem, 12, 2017
|
|
4OOU
 
 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-1,4-mannanase | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
6BOR
 
 | | Human APE1 substrate complex with an G/G mismatch adjacent the THF | | Descriptor: | 1,2-ETHANEDIOL, 21-mer DNA, CHLORIDE ION, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4TU1
 
 | | Structure of Toxoplasma gondii fructose 1,6 bisphosphate aldolase | | Descriptor: | Fructose-1,6-bisphosphate aldolase, GLYCEROL | | Authors: | Boucher, L.E, Bosch, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-06-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Toxoplasma gondii fructose-1,6-bisphosphate aldolase.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3UMX
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indol-3-yl)methylene]-7-(azepan-1-ylmethyl)-6-hydroxybenzofuran-3(2H)-one | | Descriptor: | (2Z)-7-(azepan-1-ylmethyl)-6-hydroxy-2-(1H-indol-3-ylmethylidene)-1-benzofuran-3(2H)-one, Proto-oncogene serine/threonine-protein kinase pim-1, SULFATE ION | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6CDL
 
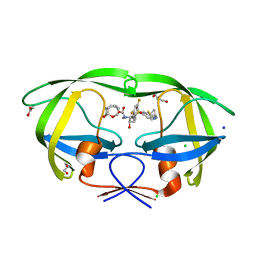 | | HIV-1 wild type protease with GRL-03214A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (2aR,4S,4aR,7aR,7bR)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
3KIZ
 
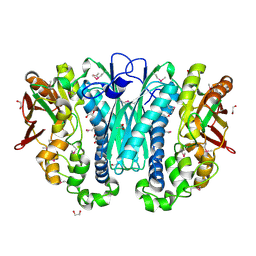 | |
5SYN
 
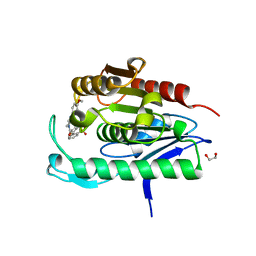 | | Cocrystal structure of the human acyl protein thioesterase 2 with an isoform-selective inhibitor, ML349 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(4-methoxyphenyl)piperazine-1-carbonyl]-5lambda~6~-thieno[3,2-c][1]benzothiopyran-5,5(4H)-dione, Acyl-protein thioesterase 2 | | Authors: | Stuckey, J.A, Labby, K.J, Meagher, J.L, Won, S.J, Martin, B.R. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2).
ACS Chem. Biol., 11, 2016
|
|
