4WBZ
 
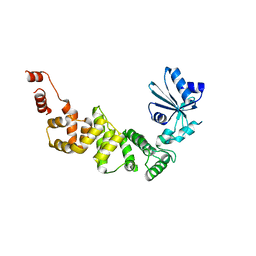 | | tRNA-processing enzyme (apo form 2) | | Descriptor: | Poly A polymerase | | Authors: | Yamashita, S, Tomtia, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
4WCA
 
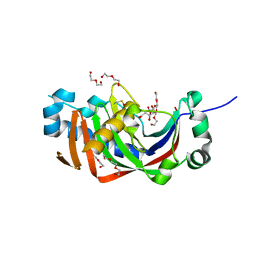 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H230Q, complexed with citrate | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
3K07
 
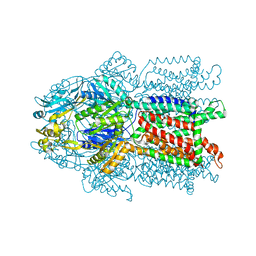 | | Crystal structure of CusA | | Descriptor: | Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
4WCZ
 
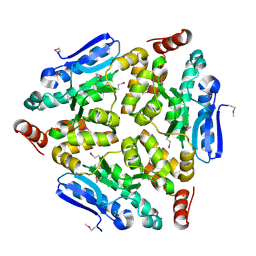 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
3K13
 
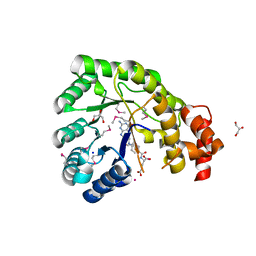 | | Structure of the pterin-binding domain MeTr of 5-methyltetrahydrofolate-homocysteine methyltransferase from Bacteroides thetaiotaomicron | | Descriptor: | 5-methyltetrahydrofolate-homocysteine methyltransferase, GLYCEROL, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, ... | | Authors: | Cuff, M.E, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-25 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the pterin-binding domain MeTr of 5-methyltetrahydrofolate-homocysteine methyltransferase from Bacteroides thetaiotaomicron
TO BE PUBLISHED
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
3K21
 
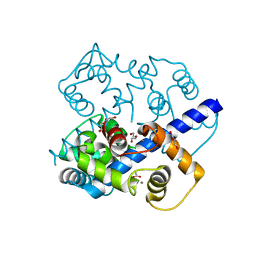 | | Crystal Structure of carboxy-terminus of PFC0420w. | | Descriptor: | ACETATE ION, CALCIUM ION, Calcium-dependent protein kinase 3, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Artz, J.D, Mackenzie, F, Cossar, D, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of parasitic CDPK domains point to a common mechanism of activation.
Proteins, 79, 2011
|
|
3K2A
 
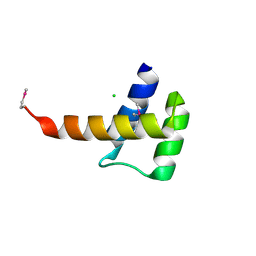 | | Crystal structure of the homeobox domain of human homeobox protein Meis2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Homeobox protein Meis2 | | Authors: | Lam, R, Soloveychik, M, Battaile, K.P, Romanov, V, Lam, K, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the homeobox domain of human homeobox protein Meis2
To be Published
|
|
4V40
 
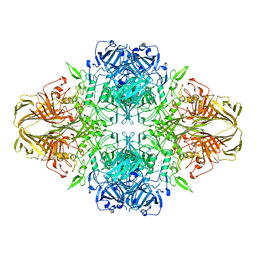 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
3S4D
 
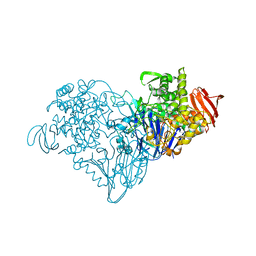 | | Lactose phosphorylase in a ternary complex with cellobiose and sulfate | | Descriptor: | Lactose Phosphorylase, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Hoorebeke, A, Stout, J, Soetaert, W, Van Beeumen, J, Desmet, T, Savvides, S. | | Deposit date: | 2011-05-19 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cellobiose phosphorylase: reconstructing the structural itinerary along the catalytic pathway
To be Published
|
|
3K4Y
 
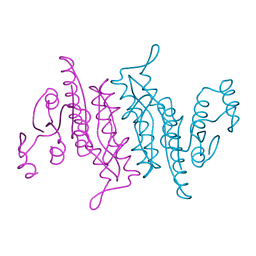 | |
3K56
 
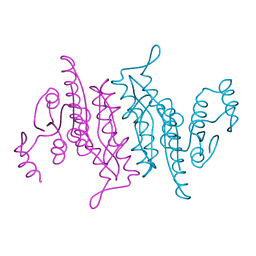 | |
3K5F
 
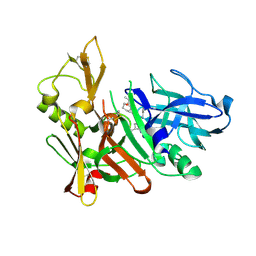 | | Human BACE-1 COMPLEX WITH AYH011 | | Descriptor: | (1R,3S)-3-[1-(acetylamino)-1-methylethyl]-N-[(1S,2S,4R)-1-benzyl-5-(butylamino)-2-hydroxy-4-methyl-5-oxopentyl]cyclohexanecarboxamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3K6N
 
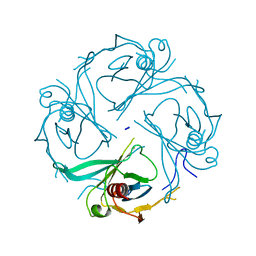 | | Crystal structure of the S225E mutant Kir3.1 cytoplasmic pore domain | | Descriptor: | G protein-activated inward rectifier potassium channel 1, SODIUM ION | | Authors: | Xu, Y, Shin, H.G, Szep, S, Lu, Z. | | Deposit date: | 2009-10-09 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Physical determinants of strong voltage sensitivity of K(+) channel block.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3K6U
 
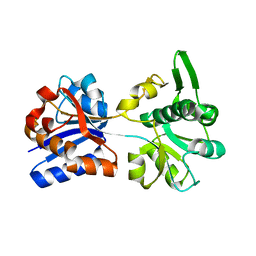 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3JQ0
 
 | |
3SI1
 
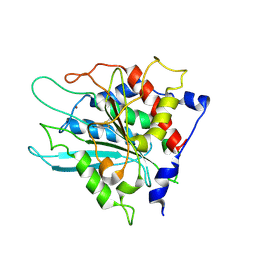 | | Structure of glycosylated murine glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Dambe, T, Carrillo, D, Parthier, C, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
3K74
 
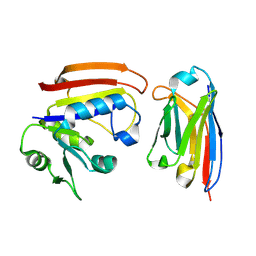 | | Disruption of protein dynamics by an allosteric effector antibody | | Descriptor: | Dihydrofolate reductase, Nanobody | | Authors: | Oyen, D, Srinivasan, V, Steyaert, J, Barlow, J. | | Deposit date: | 2009-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
J.Mol.Biol., 407, 2011
|
|
3K82
 
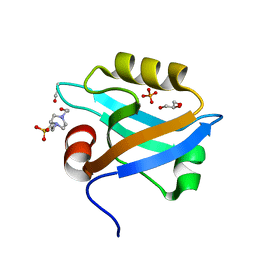 | | Crystal Structure of the third PDZ domain of PSD-95 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disks large homolog 4, GLYCEROL, ... | | Authors: | Camara-Artigas, A, Gavira, J.A. | | Deposit date: | 2009-10-13 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel conformational aspects of the third PDZ domain of the neuronal post-synaptic density-95 protein revealed from two 1.4A X-ray structures
J.Struct.Biol., 170, 2010
|
|
3K8A
 
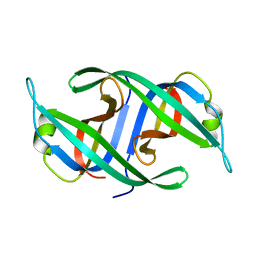 | | Neisseria gonorrhoeae PriB | | Descriptor: | Putative primosomal replication protein | | Authors: | Lopper, M.E, Dong, J, George, N.P, Duckett, K.L, DeBeer, M.A. | | Deposit date: | 2009-10-14 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Neisseria gonorrhoeae PriB reveals mechanistic differences among bacterial DNA replication restart pathways
Nucleic Acids Res., 38, 2010
|
|
3KA8
 
 | | Frog M-ferritin, EQH mutant, with cobalt | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferritin, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-19 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites.
J.Am.Chem.Soc., 132, 2010
|
|
3JRE
 
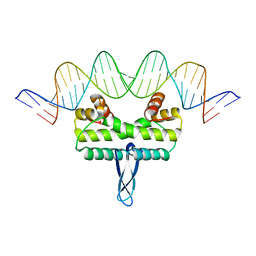 | |
3JSA
 
 | | Homoserine dehydrogenase from Thermoplasma volcanium complexed with NAD | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Osipiuk, J, Nocek, B, Hendricks, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of Homoserine dehydrogenase from Thermoplasma volcanium
To be Published
|
|
3K14
 
 | | Co-crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 535, ethyl 3-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole-2-carboxylate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
3K3K
 
 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
