6YPH
 
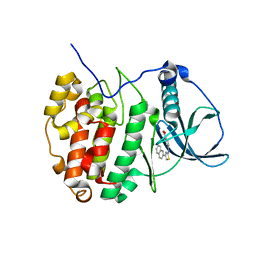 | | Crystal Structure of CK2alpha with Compound 2 bound | | Descriptor: | 4-[(4-naphthalen-2-yl-1,3-thiazol-2-yl)amino]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
3F6L
 
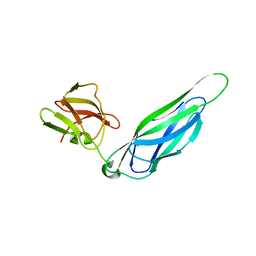 | | Structure of the F4 fimbrial chaperone FaeE | | Descriptor: | Chaperone protein faeE | | Authors: | Van Molle, I, Moonens, K, Buts, L, Garcia-Pino, A, Wyns, L, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | The F4 fimbrial chaperone FaeE is stable as a monomer that does not require self-capping of its pilin-interactive surfaces
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6YGB
 
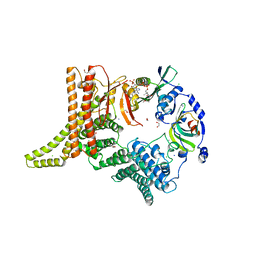 | | Crystal structure of the NatC complex bound to CoA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6P4C
 
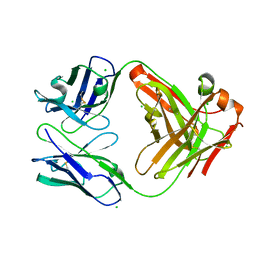 | | HyHEL10 Fab carrying four heavy chain mutations (HyHEL10-4x): L4F, Y33H, S56N, and Y58F | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3O74
 
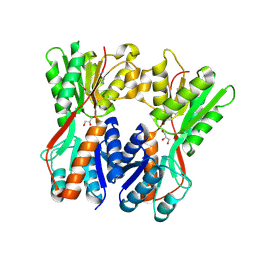 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida | | Descriptor: | Fructose transport system repressor FruR, GLYCEROL | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3O7S
 
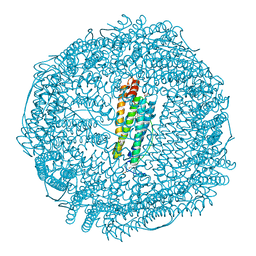 | | Crystal structure of Ru(p-cymene)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-(1-methylethyl)benzene, CADMIUM ION, ... | | Authors: | Takezawa, Y, Bockmann, P, Sugi, N, Wang, Z, Abe, S, Murakami, T, Hikage, T, Erker, G, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-07-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Incorporation of organometallic Ru complexes into apo-ferritin cage.
J.Chem.Soc.,Dalton Trans., 40, 2011
|
|
3FK0
 
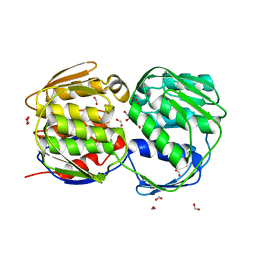 | | E. coli EPSP synthase (TIPS mutation) liganded with S3P | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Schonbrunn, E. | | Deposit date: | 2008-12-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Glyphosate Resistance Resulting from the Double Mutation Thr97 -> Ile and Pro101 -> Ser in 5-Enolpyruvylshikimate-3-phosphate Synthase from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3F6Q
 
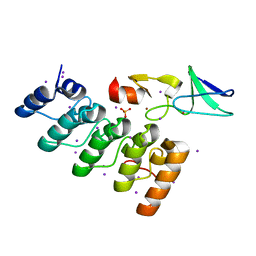 | | Crystal structure of integrin-linked kinase ankyrin repeat domain in complex with PINCH1 LIM1 domain | | Descriptor: | IODIDE ION, Integrin-linked protein kinase, LIM and senescent cell antigen-like-containing domain protein 1, ... | | Authors: | Chiswell, B.P, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2008-11-06 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis of integrin-linked kinase-PINCH interactions.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6YPT
 
 | |
3NRC
 
 | | Crystal Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD+ and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase (NADH), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Mehboob, S, Santarsiero, B.D, Truong, K, Johnson, M.E. | | Deposit date: | 2010-06-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of the Francisella tularensis enoyl-acyl carrier protein reductase (FabI) in complex with NAD(+) and triclosan.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3EX7
 
 | |
3OAA
 
 | |
7Q8T
 
 | | Crystal structure of NAMPT bound to ligand TSY535(compound 9a) | | Descriptor: | Nicotinamide phosphoribosyltransferase, SULFATE ION, [(2~{R},3~{S},4~{R},5~{S})-3,4-bis(oxidanyl)-5-[4-[[[4-(phenylsulfonyl)phenyl]carbamoylamino]methyl]phenyl]oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Kraemer, A, Tang, S, Butterworth, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Chemistry-led investigations into the mode of action of NAMPT activators, resulting in the discovery of non-pyridyl class NAMPT activators.
Acta Pharm Sin B, 13, 2023
|
|
6YLO
 
 | | mTurquoise2 - Directionality of Optical Properties of Fluorescent Proteins | | Descriptor: | POTASSIUM ION, TETRAETHYLENE GLYCOL, mTurquoise2_C2221 | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3NWI
 
 | |
6YDJ
 
 | | P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with glucose 6-phosphate and trifluoromagnesate | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
6P4D
 
 | | Hen egg lysozyme (HEL) containing three point mutations (HEL3x): R21Q, R73E, and D101R | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6YDK
 
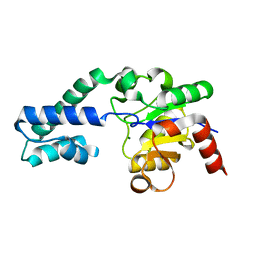 | | Substrate-free P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
3F0Q
 
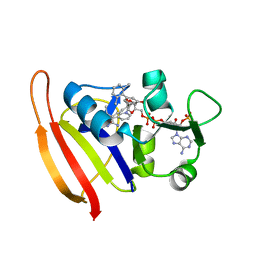 | | Staphylococcus aureus dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(2,6-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3S)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Predicting resistance mutations using protein design algorithms.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7QGJ
 
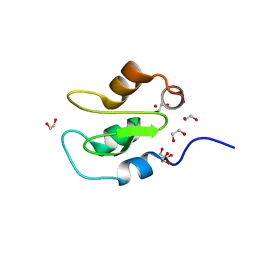 | | Apo structure of BIR2 Domain of BIRC2 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Kraemer, A, Farges, F, Schwalm, M.P, Saxena, K, Preuss, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Apo structure BIR2 Domain of BIRC2
To Be Published
|
|
3OAY
 
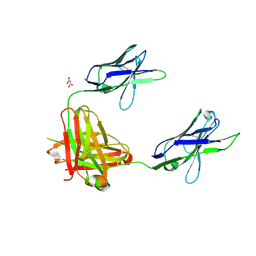 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OBR
 
 | |
3EYH
 
 | | Crystal structures of JAK1 and JAK2 inhibitor complexes | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Tyrosine-protein kinase | | Authors: | Williams, N.K, Bamert, R.S, Patell, O, Wang, C, Walden, P.M, Fantino, E. | | Deposit date: | 2008-10-20 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains.
J.Mol.Biol., 387, 2009
|
|
6YPK
 
 | | Crystal Structure of CK2alpha with GTP bound | | Descriptor: | Casein kinase II subunit alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
6YHK
 
 | | Crystal structure of full-length CNFy (C866S) from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, Cytotoxic necrotizing factor, SULFATE ION | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
