1JO4
 
 | |
2M6A
 
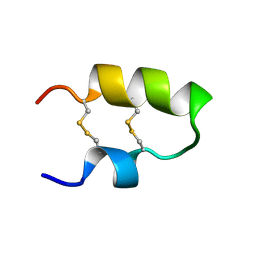 | | NMR spatial structure of the antimicrobial peptide Tk-Amp-X2 | | Descriptor: | Predicted protein | | Authors: | Usmanova, D.R, Mineev, K.S, Arseniev, A.S, Berkut, A.A, Oparin, P.B, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design
J.Biol.Chem., 289, 2014
|
|
1BRD
 
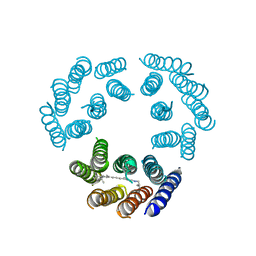 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
8BF9
 
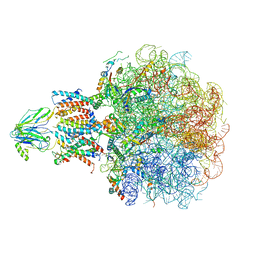 | | Molecular view of ER membrane remodeling by the Sec61/TRAP translocon. | | Descriptor: | 60S ribosomal protein L39, Large ribosomal subunit protein eL31, Large ribosomal subunit protein eL38, ... | | Authors: | Karki, S, Javanainen, M, Tranter, D, Rehan, S, Huiskonen, J, Happonen, L, Paavilainen, V. | | Deposit date: | 2022-10-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Molecular view of ER membrane remodeling by the Sec61/TRAP translocon.
Embo Rep., 24, 2023
|
|
4WNT
 
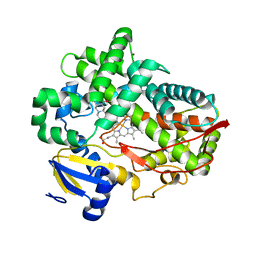 | | Human Cytochrome P450 2D6 Ajmalicine Complex | | Descriptor: | Ajmalicine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WNW
 
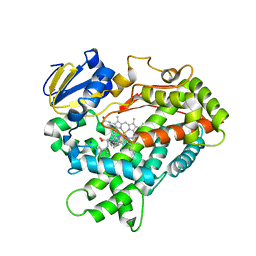 | | Human Cytochrome P450 2D6 Thioridazine Complex | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Cytochrome P450 2D6, NICKEL (II) ION, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WNV
 
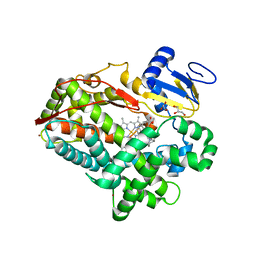 | | Human Cytochrome P450 2D6 Quinine Complex | | Descriptor: | Cytochrome P450 2D6, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WNU
 
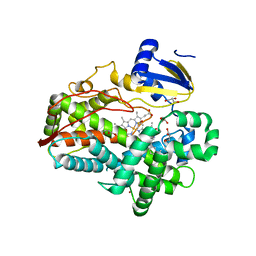 | | Human Cytochrome P450 2D6 Quinidine Complex | | Descriptor: | Cytochrome P450 2D6, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
7WR5
 
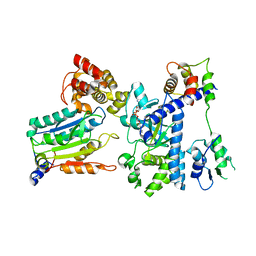 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6Z3T
 
 | | Structure of canine Sec61 inhibited by mycolactone | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Gerard, S.F, Higgins, M.K. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Inhibited State of the Sec Translocon.
Mol.Cell, 79, 2020
|
|
6QS8
 
 | |
6QS4
 
 | |
6QS7
 
 | |
6QS6
 
 | |
3LOY
 
 | | Crystal structure of a Copper-containing benzylamine oxidase from Hansenula Polymorpha | | Descriptor: | COPPER (II) ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Klema, V.J, Johnson, B.J, Wilmot, C.M. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of substrate specificity in two copper amine oxidases from Hansenula polymorpha.
Biochemistry, 49, 2010
|
|
8OF8
 
 | | Cryo-EM structure of actomyosin-5a-S1 with the full-length lever (nucleotide free) | | Descriptor: | Actin, alpha skeletal muscle, Calmodulin-1, ... | | Authors: | Gravett, M.S.C, Klebl, D.P, Harlen, O.G, Read, D.J, Harris, S.A, Muench, S.P, Peckham, M. | | Deposit date: | 2023-03-14 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Unveiling the Structural Dynamics of Myosin 5a: Cryo-EM Insights into the Motor Protein's Lever Conformations and Walking Mechanics
To Be Published
|
|
2WEL
 
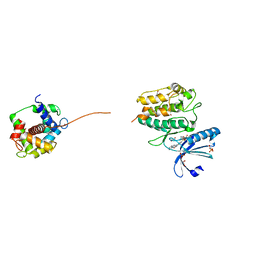 | | Crystal structure of SU6656-bound calcium/calmodulin-dependent protein kinase II delta in complex with calmodulin | | Descriptor: | (3Z)-N,N-DIMETHYL-2-OXO-3-(4,5,6,7-TETRAHYDRO-1H-INDOL-2-YLMETHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-5-SULFONAMIDE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Salah, E, Burgess-Brown, N, Keates, T, Muniz, J, Sethi, R, Roos, A, Filippakopoulos, P, von Delft, F, Edwards, A, Weigelt, J, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Camkiidelta/Calmodulin Complex Reveals the Molecular Mechanism of Camkii Kinase Activation.
Plos Biol., 8, 2010
|
|
1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
1KCB
 
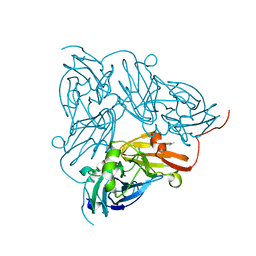 | | Crystal Structure of a NO-forming Nitrite Reductase Mutant: an Analog of a Transition State in Enzymatic Reaction | | Descriptor: | COPPER (II) ION, Nitrite Reductase | | Authors: | Liu, S.Q, Chang, T, Liu, M.Y, LeGall, J, Chang, W.C, Zhang, J.P, Liang, D.C, Chang, W.R. | | Deposit date: | 2001-11-07 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a NO-forming nitrite reductase mutant: an analog of a transition state in enzymatic reaction
Biochem.Biophys.Res.Commun., 302, 2003
|
|
1KGY
 
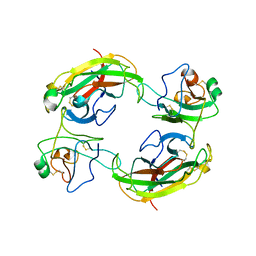 | | Crystal Structure of the EphB2-ephrinB2 complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2, EPHRIN-B2 | | Authors: | Himanen, J.P, Rajashankar, K.R, Lackmann, M, Cowan, C.A, Henkemeyer, M, Nikolov, D.B. | | Deposit date: | 2001-11-28 | | Release date: | 2002-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an Eph receptor-ephrin complex.
Nature, 414, 2001
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
1WYW
 
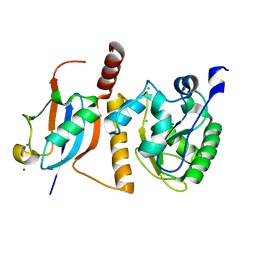 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
1A5R
 
 | | STRUCTURE DETERMINATION OF THE SMALL UBIQUITIN-RELATED MODIFIER SUMO-1, NMR, 10 STRUCTURES | | Descriptor: | SUMO-1 | | Authors: | Bayer, P, Arndt, A, Metzger, S, Mahajan, R, Melchior, F, Jaenicke, R, Becker, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the small ubiquitin-related modifier SUMO-1.
J.Mol.Biol., 280, 1998
|
|
2PE6
 
 | | Non-covalent complex between human SUMO-1 and human Ubc9 | | Descriptor: | SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Capili, A.D, Lima, C.D. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Analysis of a Complex between SUMO and Ubc9 Illustrates Features of a Conserved E2-Ubl Interaction.
J.Mol.Biol., 369, 2007
|
|
3ROW
 
 | | Crystal Structure of Xanthomonas campestri OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III | | Authors: | Goblirsch, B.R, Wilmot, C.M. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8487 Å) | | Cite: | Crystal Structures of Xanthomonas campestris OleA Reveal Features That Promote Head-to-Head Condensation of Two Long-Chain Fatty Acids.
Biochemistry, 51, 2012
|
|
