6IZ4
 
 | | Crystal Structure Analysis of TRIC counter-ion channels in calcium release | | Descriptor: | Trimeric intracellular cation channel type B-B | | Authors: | Wang, X.H, Zeng, Y, Gao, F, Su, M, Hendrickson, W.A, Chen, Y.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7PVY
 
 | |
6FL8
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with purpurogallin and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6PNB
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(4-(Aminomethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[4-(aminomethyl)phenyl]-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7ZND
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.6 in the absence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5UG7
 
 | | Calcium bound Perforin C2 Domain - T431D | | Descriptor: | CALCIUM ION, Perforin-1 | | Authors: | Law, R.H.P, Conroy, P.J, Voskoboinik, I, Whisstock, J.C. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Perforin proteostasis is regulated through its C2 domain: supra-physiological cell death mediated by T431D-perforin.
Cell Death Differ., 25, 2018
|
|
7KV7
 
 | | Surface glycan-binding protein B from Bacteroides fluxus in complex with laminaritriose | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, LYSINE, ... | | Authors: | Tamura, K, Brumer, H, Van Petegem, F. | | Deposit date: | 2020-11-26 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Distinct protein architectures mediate species-specific beta-glucan binding and metabolism in the human gut microbiota.
J.Biol.Chem., 296, 2021
|
|
7ZQ2
 
 | | Crystal structure of Pizza6-RSH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-RSH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
6YJC
 
 | | Crystal structure of p38alpha in complex with SR154 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-oxidanylidene-1-piperazin-1-yl-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74100935 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
5UGI
 
 | |
6F3Z
 
 | | Complex of E. coli LolA and periplasmic domain of LolC | | Descriptor: | Lipoprotein-releasing system transmembrane protein LolC, Outer-membrane lipoprotein carrier protein | | Authors: | Kaplan, E. | | Deposit date: | 2017-11-29 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7ZN8
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PNO
 
 | |
5UGR
 
 | | Malyl-CoA lyase from Methylobacterium extorquens | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Malyl-CoA lyase/beta-methylmalyl-CoA lyase, ... | | Authors: | Gonzalez, J.M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Methylobacterium extorquens malyl-CoA lyase: CoA-substrate binding correlates with domain shift.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7L07
 
 | | Last common ancestor of HMPPK and PLK/HMPPK vitamin kinases | | Descriptor: | ALUMINUM FLUORIDE, Ancestral Protein AncC | | Authors: | Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Araya, G, Arizabalos, S, Castro-Fernandez, V. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and molecular dynamics simulations of a promiscuous ancestor reveal residues and an epistatic interaction involved in substrate binding and catalysis in the ATP-dependent vitamin kinase family members.
Protein Sci., 30, 2021
|
|
7ZQG
 
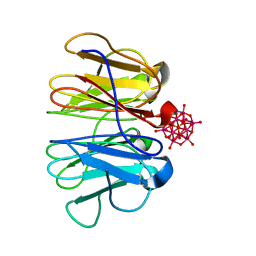 | | Crystal structure of Pizza6-KSH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-KSH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
6B85
 
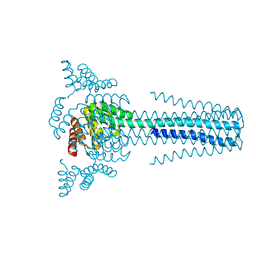 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6J0O
 
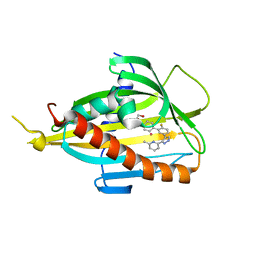 | | Crystal structure of CERT START domain in complex with compound SC1 | | Descriptor: | 2-[4-[2-fluoranyl-5-[3-(6-methylpyridin-2-yl)-1~{H}-pyrazol-4-yl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT, UNKNOWN ATOM OR ION | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2018-12-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
6YHM
 
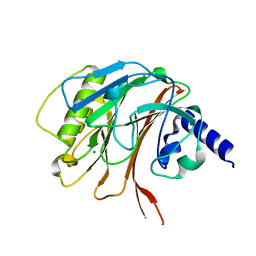 | | Crystal structure of the C-terminal domain of CNFy from Yersinia pseudotuberculosis | | Descriptor: | Cytotoxic necrotizing factor, MAGNESIUM ION | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
7PQI
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7KV5
 
 | |
6PMV
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(4-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[4-(2-aminoethyl)phenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7ZNC
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 7.6 in the absence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5FIY
 
 | | crystal structure of coiled coil domain of PAWR | | Descriptor: | POTASSIUM ION, PRKC APOPTOSIS WT1 REGULATOR PROTEIN | | Authors: | Tiruttani Subhramanyam, U.K, Kubicek, J, Eidhoff, U.B, Labahn, J. | | Deposit date: | 2015-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the regulatory interactions of proapoptotic Par-4.
Cell Death Differ., 24, 2017
|
|
6J0X
 
 | | Crystal Structure of Yeast Rtt107 and Mms22 | | Descriptor: | Peptide from E3 ubiquitin-protein ligase substrate receptor MMS22, Regulator of Ty1 transposition protein 107 | | Authors: | Wan, B, Wu, J, Lei, M. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
