5VYY
 
 | |
5DGA
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA EXTENDING AN 1,N6-ETHENODEOXYADENOSINE : dT PAIR BY INSERTING dTMPNPP OPPOSITE TEMPLATE dA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*AP*(EDA)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
6LTU
 
 | | Crystal structure of Cas12i2 ternary complex with double Mg2+ bound in catalytic pocket | | Descriptor: | 1,2-ETHANEDIOL, Cas12i2, DNA (35-MER), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
5VCR
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound uranium dioxide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, CHLORIDE ION, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CLU
 
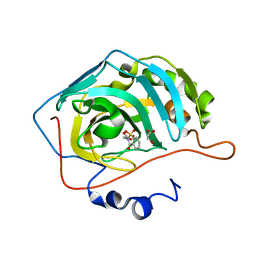 | | THE CRYSTAL STRUCTURE OF THE COMPLEX of HCAII WITH A SACCHARINE DERIVATIVE | | Descriptor: | (1,1-dioxido-3-oxo-1,2-benzothiazol-2(3H)-yl)acetic acid, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | D'Ambrosio, K, De Simone, G. | | Deposit date: | 2015-07-16 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Combined Crystallographic and Theoretical Study Explains the Capability of Carboxylic Acids to Adopt Multiple Binding Modes in the Active Site of Carbonic Anhydrases.
Chemistry, 22, 2016
|
|
7Z28
 
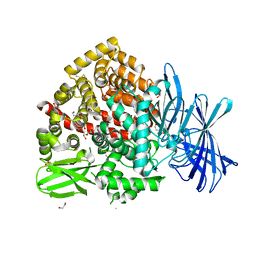 | | High-resolution crystal structure of ERAP1 with bound bestatin analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Papakyriakou, A, Stratikos, E, Vourloumis, D. | | Deposit date: | 2022-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
6B5G
 
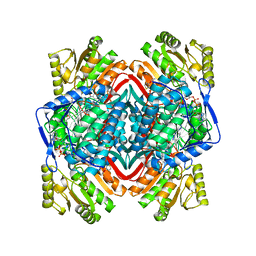 | |
4BHT
 
 | | Structural Determinants of Cofactor Specificity and Domain Flexibility in Bacterial Glutamate Dehydrogenases | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NADP-SPECIFIC GLUTAMATE DEHYDROGENASE, ... | | Authors: | Oliveira, T, Sharkey, M, Engel, P, Khan, A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nadp(+) -Dependent Glutamate Dehydrogenase from Escherichia Coli: Reflections on the Basis of Coenzyme Specificity in the Family of Glutamate Dehydrogenases.
FEBS J., 280, 2013
|
|
6N1T
 
 | | Toxoplasma gondii TS-DHFR in complex with selective inhibitor 3 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-{4-[3-(2-methoxypyrimidin-5-yl)phenyl]piperazin-1-yl}pyrimidine-2,4-diamine, ... | | Authors: | Hopper, A.T, Brockman, A, Wise, A, Gould, J, Barks, J, Radke, J.B, Sibley, L.D, Zou, Y, Thomas, S.B. | | Deposit date: | 2018-11-11 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of Selective Toxoplasma gondii Dihydrofolate Reductase Inhibitors for the Treatment of Toxoplasmosis.
J. Med. Chem., 62, 2019
|
|
4O5H
 
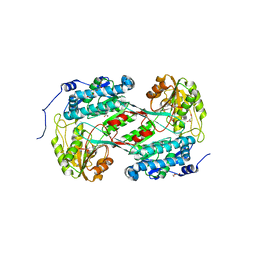 | |
2WCW
 
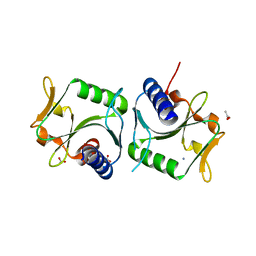 | | 1.6A resolution structure of Archaeoglobus fulgidus Hjc, a Holliday junction resolvase from an archaeal hyperthermophile | | Descriptor: | ACETATE ION, AMMONIUM ION, HJC | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | 1.6 A Resolution Structure of Archaeoglobus Fulgidus Hjc, a Holliday Junction Resolvase from an Archaeal Hyperthermophile
To be Published
|
|
6MAZ
 
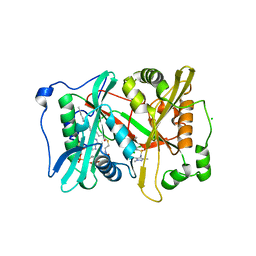 | |
6NAU
 
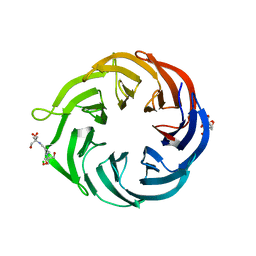 | | 1.55 Angstrom Resolution Crystal Structure of 6-phosphogluconolactonase from Klebsiella pneumoniae | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-phosphogluconolactonase, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Dubrovska, I, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6CEO
 
 | |
6MQA
 
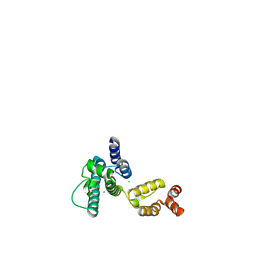 | | Structure of HIV-1 CA P207S | | Descriptor: | CHLORIDE ION, Capsid protein, IODIDE ION | | Authors: | Smaga, S.S, Xiong, Y. | | Deposit date: | 2018-10-09 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | MxB Restricts HIV-1 by Targeting the Tri-hexamer Interface of the Viral Capsid.
Structure, 27, 2019
|
|
6CIS
 
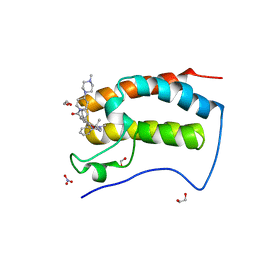 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG047 | | Descriptor: | 1,2-ETHANEDIOL, 11-cyclopentyl-5-methyl-2-({4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]-2-[(propan-2-yl)oxy]phenyl}amino)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
6MVY
 
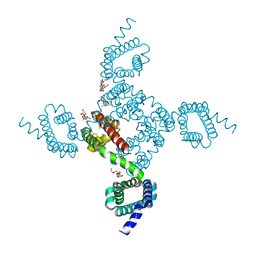 | | NavAb voltage-gated sodium channel, residues 1-226, crystallized in the presence of Class 1B Anti-arrhythmic drug Lidocaine | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein, SULFATE ION | | Authors: | Lenaeus, M.J, Catterall, W.A. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Fenestrations control resting-state block of a voltage-gated sodium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3WKT
 
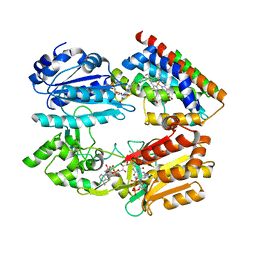 | | Complex structure of an open form of NADPH-cytochrome P450 reductase and heme oxygenase-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Sato, H, Higashimoto, Y, Harada, J, Wada, K, Fukuyama, K, Noguchi, M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CD4
 
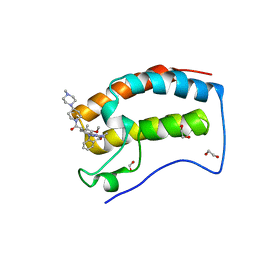 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG046 | | Descriptor: | 1,2-ETHANEDIOL, 2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-11-(propan-2-yl)-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-08 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
3WQS
 
 | | Crystal structure of pfdxr complexed with inhibitor-126 | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, MAGNESIUM ION, ... | | Authors: | Tanaka, N, Umeda, T. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Binding Modes of Reverse Fosmidomycin Analogs toward the Antimalarial Target IspC.
J.Med.Chem., 57, 2014
|
|
7C5K
 
 | | Crystal Structure of C150S mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with G3P at 2.69 Angstrom resolution | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
2QG4
 
 | | Crystal structure of human UDP-glucose dehydrogenase product complex with UDP-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kavanagh, K.L, Guo, K, Bunkoczi, G, Savitsky, P, Pilka, E, Bhatia, C, Niesen, F, Smee, C, Berridge, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
3URH
 
 | | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | 1,2-ETHANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021
TO BE PUBLISHED
|
|
3IMF
 
 | | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
7LD8
 
 | |
