6TXJ
 
 | | Crystal structure of thermotoga maritima A42V E65D Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S, Biela, A.P. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXL
 
 | | Crystal structure of thermotoga maritima E65Q Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXK
 
 | | Crystal structure of thermotoga maritima E65K Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXI
 
 | | Crystal structure of thermotoga maritima E65A Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6U46
 
 | |
6UCO
 
 | |
6UCP
 
 | |
4WO9
 
 | | Lysozyme Post-Surface Acoustic Waves | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WHA
 
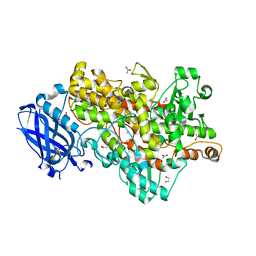 | | Lipoxygenase-1 (soybean) L546A/L754A mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Scouras, A.D, Carr, C.A.M, Hu, S, Klinman, J.P. | | Deposit date: | 2014-09-21 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extremely elevated room-temperature kinetic isotope effects quantify the critical role of barrier width in enzymatic C-H activation.
J.Am.Chem.Soc., 136, 2014
|
|
6UQK
 
 | | Cryo-EM structure of type 3 IP3 receptor revealing presence of a self-binding peptide | | Descriptor: | ZINC ION, inositol 1,4,5-triphosphate receptor, type 3 | | Authors: | Azumaya, C.M, Linton, E.A, Risener, C.J, Nakagawa, T, Karakas, E. | | Deposit date: | 2019-10-20 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structure of human type-3 inositol triphosphate receptor reveals the presence of a self-binding peptide that acts as an antagonist.
J.Biol.Chem., 295, 2020
|
|
4WGX
 
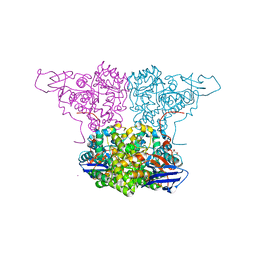 | | Crystal Structure of Molinate Hydrolase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COBALT (II) ION, Molinate hydrolase | | Authors: | Sugrue, E, Carr, P.D, Fraser, N.J, Hopkins, D.H, Jackson, C.J. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
7AJU
 
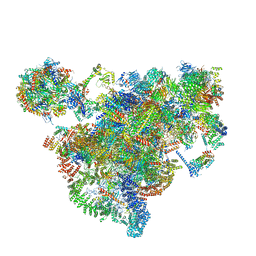 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
4WH3
 
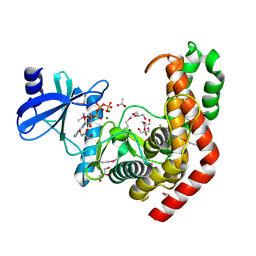 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WL6
 
 | | Raster-scanning protein crystallography using micro and nano-focused synchrotron beams | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Kapp, U, Shilova, A, Weinhausen, B, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-10-06 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7AJT
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
8E31
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | Genome polyprotein, VP1, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
4X5H
 
 | | ecDHFR complexed with folate and NADP+ at 500 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Yamada, H, Watanabe, N, Nagae, T. | | Deposit date: | 2014-12-05 | | Release date: | 2016-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-pressure protein crystal structure analysis of Escherichia coli dihydrofolate reductase complexed with folate and NADP.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4V7Z
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with telithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7TUJ
 
 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-09 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
4WOB
 
 | | Proteinase-K Pre-Surface Acoustic Wave | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WO6
 
 | | Lysozyme Pre-surface acoustic wave | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | French, J.B. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Precise Manipulation and Patterning of Protein Crystals for Macromolecular Crystallography Using Surface Acoustic Waves.
Small, 11, 2015
|
|
4WVF
 
 | | Crystal structure of KPT276 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2E)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-1-(3,3-difluoroazetidin-1-yl)prop-2-en-1-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Chook, Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination.
Nat.Neurosci., 18, 2015
|
|
6PQM
 
 | |
4X5G
 
 | | ecDHFR complexed with folate and NADP+ at 270 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Yamada, H, Watanabe, N, Nagae, T. | | Deposit date: | 2014-12-05 | | Release date: | 2016-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-pressure protein crystal structure analysis of Escherichia coli dihydrofolate reductase complexed with folate and NADP.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4X5J
 
 | | ecDHFR complexed with folate and NADP+ at 750 MPa | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Yamada, H, Watanabe, N, Nagae, T. | | Deposit date: | 2014-12-05 | | Release date: | 2016-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-pressure protein crystal structure analysis of Escherichia coli dihydrofolate reductase complexed with folate and NADP.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
