6G67
 
 | |
7D3I
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with MI-23 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[3-[3,5-bis(fluoranyl)phenyl]propanoyl]-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase | | Authors: | Zeng, R, Li, Y.S, Qiao, J.X, Wang, Y.F, Yang, S.Y, Lei, J. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
6G6A
 
 | |
5TWZ
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
8OZS
 
 | | Populus tremula stable protein 1 with N-terminal binding peptide extension with hemin | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2023
|
|
6FRV
 
 | | Structure of the catalytic domain of Aspergillus niger Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucoamylase, ... | | Authors: | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2018-02-16 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7DC8
 
 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | Authors: | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
5DSG
 
 | | Structure of the M4 muscarinic acetylcholine receptor (M4-mT4L) bound to tiotropium | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Thal, D.M, Kobilka, B.K, Sexton, P.M, Christopoulos, A. | | Deposit date: | 2015-09-17 | | Release date: | 2016-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the M1 and M4 muscarinic acetylcholine receptors.
Nature, 531, 2016
|
|
6G6D
 
 | |
6VFW
 
 | | Crystal structure of human delta protocadherin 10 EC1-EC4 | | Descriptor: | CALCIUM ION, Protocadherin-10, alpha-D-mannopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
8OZO
 
 | | Populus tremula stable protein 1 with N-terminal binding peptide extension | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2024
|
|
6G6G
 
 | |
5L6I
 
 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
6FSL
 
 | |
5TXY
 
 | | Identification of a New Zinc Binding Chemotype of by Fragment Screening on human carbonic anhydrase | | Descriptor: | (5R)-5-phenyl-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B, Peat, T.S, Poulsen, S.-A. | | Deposit date: | 2016-11-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5HWP
 
 | | MvaS with acetylated Cys115 in complex with coenzyme A | | Descriptor: | COENZYME A, GLYCEROL, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Bock, T, Kasten, J, Blankenfeldt, W. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-11 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the HMG-CoA Synthase MvaS from the Gram-Negative Bacterium Myxococcus xanthus.
Chembiochem, 17, 2016
|
|
6XP7
 
 | |
8ODO
 
 | | Structure of human guanylylated RTCB in complex with Archease | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
5TYA
 
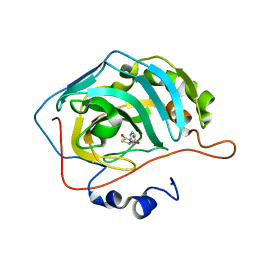 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-phenyl-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
6MNZ
 
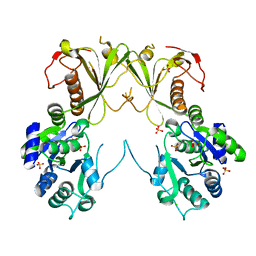 | | Crystal structure of RibBX, a two domain 3,4-dihydroxy-2-butanone 4-phosphate synthase from A. baumannii. | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Wang, J, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Multi-metal Restriction by Calprotectin Impacts De Novo Flavin Biosynthesis in Acinetobacter baumannii.
Cell Chem Biol, 26, 2019
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6G6M
 
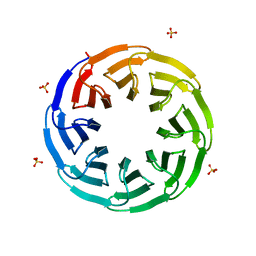 | | Crystal structure of the computationally designed Tako8 protein in P42212 | | Descriptor: | SULFATE ION, Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
5L6W
 
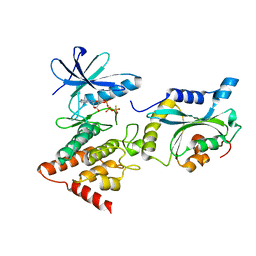 | | Structure Of the LIMK1-ATPgammaS-CFL1 Complex | | Descriptor: | Cofilin-1, LIM domain kinase 1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Salah, E, Mathea, S, Oerum, S, Newman, J.A, Tallant, C, Adamson, R, Canning, P, Beltrami, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Bullock, A.N. | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure Of the LIMK1-ATPgammaS-CFL1 Complex
To Be Published
|
|
6XPC
 
 | | Structure of human GGT1 in complex with full GSH molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of glutathione- and inhibitor-bound human GGT1: critical interactions within the cysteinylglycine binding site.
J.Biol.Chem., 296, 2020
|
|
6VGW
 
 | | Crystal structure of VidaL intein (selenomethionine variant) | | Descriptor: | GLYCEROL, SULFATE ION, VidaL | | Authors: | Burton, A.J, Haugbro, M, Parisi, E, Muir, T.W. | | Deposit date: | 2020-01-09 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Live-cell protein engineering with an ultra-short split intein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
