8A3X
 
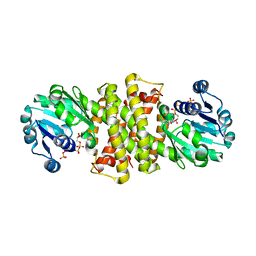 | | Imine Reductase from Ensifer adhaerens in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NAD_binding_2 domain-containing protein, POTASSIUM ION | | Authors: | Gilio, A.K, Grogan, G. | | Deposit date: | 2022-06-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Reductive Aminase Switches to Imine Reductase Mode for a Bulky Amine Substrate.
Acs Catalysis, 13, 2023
|
|
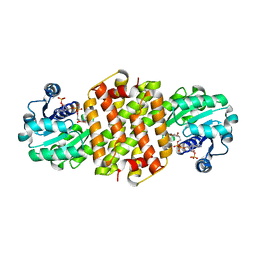
8BJ5
 
 | |
1H8A
 
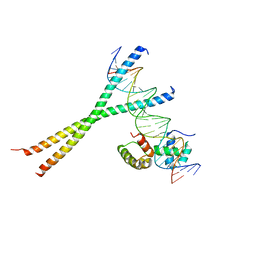 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-31 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1H89
 
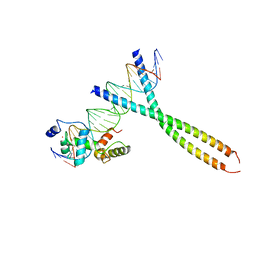 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-30 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
6D9G
 
 | |
7UE1
 
 | | HIV-1 Integrase Catalytic Core Domain Mutant (KGD) in Complex with Inhibitor GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Integrase, SULFATE ION | | Authors: | Aoki, M, Aoki-Ogata, H, Bulut, H, Hayashi, H, Davis, D, Hasegawa, K, Yarchoan, R, Ghosh, A.K, Pau, A.K, Mitsuya, H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants.
Sci Adv, 9, 2023
|
|
5VHB
 
 | |
5VIB
 
 | |
5VI9
 
 | |
6OYR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-002 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-1-(3-fluorophenyl)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]butan-2-yl}carbamate, Protease | | Authors: | Bulut, H, Hattori, S.I, Aoki-Ogata, H, Hayashi, H, Aoki, M, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Sci Rep, 10, 2020
|
|
7LIM
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S6E | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIS
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S5D | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
7LIT
 
 | | Thermotoga maritima Encapsulin Nanocompartment Pore Mutant S7G | | Descriptor: | Maritimacin, RIBOFLAVIN | | Authors: | Andreas, M.P, Adamson, L, Tasneem, N, Close, W, Giessen, T, Lau, Y.H. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Pore structure controls stability and molecular flux in engineered protein cages.
Sci Adv, 8, 2022
|
|
8OZV
 
 | | Imine Reductase from Ajellomyces dermatitidis in complex with 2,2-difluoroacetophenone | | Descriptor: | 2,2-bis(fluoranyl)-1-phenyl-ethanone, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the imine reductase from Ajellomyces dermatitidis in three crystal forms.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8P23
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/CTP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
Elife, 2023
|
|
8P2J
 
 | |
8P28
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric, dATP-bound state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
To Be Published
|
|
8P27
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dATP-bound state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductases: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
Elife, 2023
|
|
8OZW
 
 | | Imine Reductase from Ajellomyces dermatitidis in complex NADPH4 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the imine reductase from Ajellomyces dermatitidis in three crystal forms.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8P2C
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric state produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P2S
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/dTTP/GTP-bound state | | Descriptor: | Anaerobic ribonucleoside-triphosphate reductase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P2D
 
 | | Cryo-EM structure of the dimeric form of the anaerobic ribonucleotide reductase from Prevotella copri produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P39
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-17 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
To be published
|
|
8PBF
 
 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-(4-phenylsulfanyl-1,2,3-triazol-1-yl)oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Mahanti, M. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
6H0T
 
 | | Crystal structure of native recombinant human bile salt activated lipase | | Descriptor: | ACETATE ION, Bile salt-activated lipase, GLYCEROL, ... | | Authors: | Touvrey, C, Brazzolotto, X, Nachon, F. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of human bile-salt activated lipase conjugated to nerve agents surrogates.
Toxicology, 411, 2019
|
|
