5KGH
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant Y297F | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KF9
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Holden, H.M, Tipton, P.A. | | Deposit date: | 2016-06-12 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGA
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, mutant D287N, in complex with N-acetylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
8DP8
 
 | |
6MH8
 
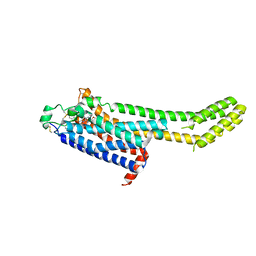 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
5L8Q
 
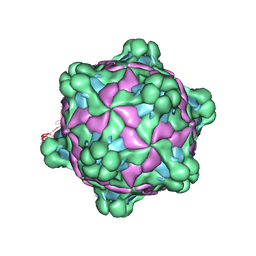 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-06-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6FQS
 
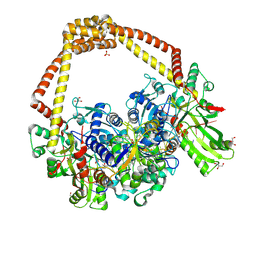 | | 3.11A complex of S.Aureus gyrase with imidazopyrazinone T3 and DNA | | Descriptor: | 5-cyclopropyl-8-fluoranyl-7-pyridin-4-yl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
7ZPG
 
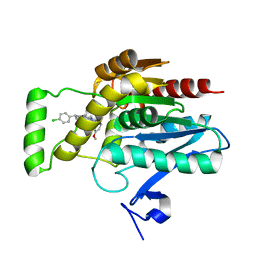 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH LIGAND | | Descriptor: | Monoglyceride lipase, [(7R,9aR)-7-(4-chlorophenyl)-1,3,4,6,7,8,9,9a-octahydropyrido[1,2-a]pyrazin-2-yl]-(2-bromanyl-3-methoxy-phenyl)methanone | | Authors: | Kemble, A, Hornsperger, B, Ruf, I, Richter, H, Benz, J, Kuhn, B, Heer, D, Wittwer, M, Engelhardt, B, Grether, U, Collin, L, Leibrock, L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A potent and selective inhibitor for the modulation of MAGL activity in the neurovasculature.
Plos One, 17, 2022
|
|
6XCL
 
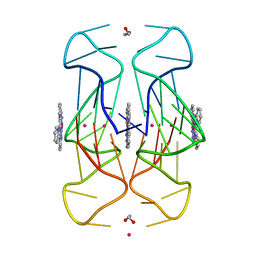 | | Crystal Structure of human telomeric DNA G-quadruplex in complex with a novel platinum(II) complex. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), POTASSIUM ION, ... | | Authors: | Miron, C.E, van Staalduinen, L.M, Jia, Z, Petitjean, A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Going Platinum to the Tune of a Remarkable Guanine Quadruplex Binder: Solution- and Solid-State Investigations.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6OCB
 
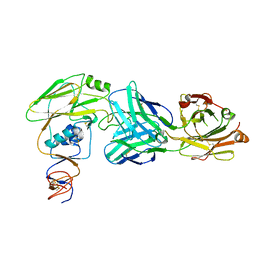 | |
5L7Q
 
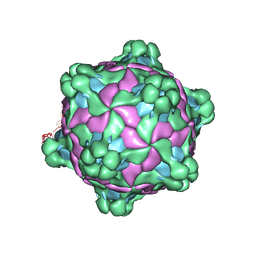 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | VP1, vp2, vp3 | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8F5Y
 
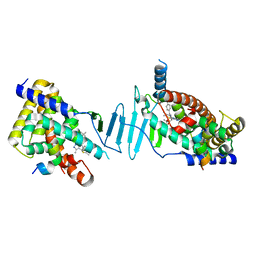 | | Crystal structure of pregnane X receptor ligand binding domain complexed with JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Nuclear receptor coactivator 1, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Chen, T. | | Deposit date: | 2022-11-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A bromodomain-independent mechanism of gene regulation by the BET inhibitor JQ1: direct activation of nuclear receptor PXR.
Nucleic Acids Res., 52, 2024
|
|
6WF3
 
 | | Crystal structure of human Naa50 in complex with a cofactor derived inhibitor (compound 1) | | Descriptor: | ACE-MET-LEU-GLY-PRO-NH2, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6H0U
 
 | | Glycogen synthase kinase-3 beta (GSK3) complex with a covalent [1,2,4]triazolo[1,5-a][1,3,5]triazine inhibitor | | Descriptor: | (2~{R})-3-[7-azanyl-5-(cyclohexylamino)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-2-yl]-2-cyano-propanamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marcovich, I, Demitri, N, De Zorzi, R, Storici, P. | | Deposit date: | 2018-07-10 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Triazolotriazine-Based Dual GSK-3 beta /CK-1 delta Ligand as a Potential Neuroprotective Agent Presenting Two Different Mechanisms of Enzymatic Inhibition.
Chemmedchem, 14, 2019
|
|
5MV6
 
 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KF1
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum, apo form, pH 5 | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Holden, H.M, Thoden, J.B, Dopkins, B.J, tipton, P.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
5KGJ
 
 | | X-ray structure of a glucosamine N-Acetyltransferase from Clostridium acetobutylicum in complex with galactosamine | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-alpha-D-galactopyranose, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, ... | | Authors: | Dopkins, B.J, Thoden, J.B, Tipton, P.A, Holden, H.M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies on a Glucosamine/Glucosaminide N-Acetyltransferase.
Biochemistry, 55, 2016
|
|
9C9L
 
 | | Cryo-EM structure of the C1q A, B-crt, C peptide assembly narrow region | | Descriptor: | Peptide from Complement C1q subcomponent subunit A, Peptide from Complement C1q subcomponent subunit B, Peptide from Complement C1q subcomponent subunit C | | Authors: | Kreutzberger, M.A, Yu, L.T, Egelman, E.H, Hartgerink, J.D. | | Deposit date: | 2024-06-14 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Collagen Triple Helix without the Superhelical Twist.
Acs Cent.Sci., 11, 2025
|
|
9C9U
 
 | | Cryo-EM structure of the C1q A, B-crt, C peptide full assembly | | Descriptor: | Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, Complement C1q subcomponent subunit C | | Authors: | Kreutzberger, M.A, Yu, L.T, Egelman, E.H, Hartgerink, J.D. | | Deposit date: | 2024-06-15 | | Release date: | 2025-02-12 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Collagen Triple Helix without the Superhelical Twist.
Acs Cent.Sci., 11, 2025
|
|
5KTC
 
 | | FdhC with bound products: Coenzyme A and 3-[(R)-3-hydroxybutanoylamino]-3,6-dideoxy-d-galactose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
4XYL
 
 | | Ca. Korarchaeum cryptofilum ACD1 in complex with coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, alpha subunit of Acyl-CoA synthetase (NDP forming), ... | | Authors: | Weisse, R.H.-J, Scheidig, A.J. | | Deposit date: | 2015-02-02 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of NDP-forming Acetyl-CoA synthetase ACD1 reveals a large rearrangement for phosphoryl transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KTD
 
 | | FdhC with bound products: Coenzyme A and dTDP-3-amino-3,6-dideoxy-d-glucose | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, FdhC, ... | | Authors: | Salinger, A.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2016-07-11 | | Release date: | 2016-07-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Investigation of FdhC from Acinetobacter nosocomialis: A Sugar N-Acyltransferase Belonging to the GNAT Superfamily.
Biochemistry, 55, 2016
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
