8JKD
 
 | |
2Q5A
 
 | | human Pin1 bound to L-PEPTIDE | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
3KFR
 
 | |
3KG1
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
8JNR
 
 | |
6IRT
 
 | | human LAT1-4F2hc complex bound with BCH | | Descriptor: | (1S,2S,4R)-2-aminobicyclo[2.2.1]heptane-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhao, X, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex.
Nature, 568, 2019
|
|
6IS0
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH and m7G-capped RNA | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
1UPT
 
 | | Structure of a complex of the golgin-245 GRIP domain with Arl1 | | Descriptor: | ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 1, GOLGI AUTOANTIGEN, GOLGIN SUBFAMILY A MEMBER 4, ... | | Authors: | Panic, B, Perisic, O, Veprintsev, D.B, Williams, R.L, Munro, S. | | Deposit date: | 2003-10-12 | | Release date: | 2003-10-30 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Arl1-Dependent Targeting of Homodimeric Grip Domains to the Golgi Apparatus
Mol.Cell, 12, 2003
|
|
6IE2
 
 | | Crystal structure of methyladenine demethylase | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
1V6R
 
 | | Solution Structure of Endothelin-1 with its C-terminal Folding | | Descriptor: | Endothelin-1 | | Authors: | Takashima, H, Mimura, N, Ohkubo, T, Yoshida, T, Tamaoki, H, Kobayashi, Y. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Distributed Computing and NMR Constraint-Based High-Resolution Structure
Determination: Applied for Bioactive Peptide Endothelin-1 To Determine C-Terminal
Folding
J.Am.Chem.Soc., 126, 2004
|
|
3KCE
 
 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 5-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8IVB
 
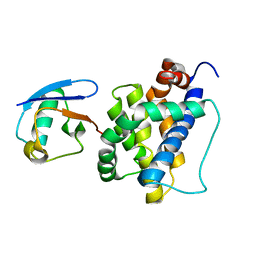 | | K113-Ubiquitinated BAK | | Descriptor: | Bcl-2 homologous antagonist/killer, Ubiquitin | | Authors: | Dong, X, Cheng, P, Hou, Y.Z, Chen, Y.K, Liu, Z. | | Deposit date: | 2023-03-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove.
Commun Biol, 6, 2023
|
|
8J9O
 
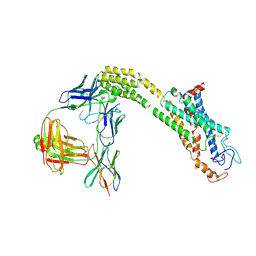 | | Cryo-EM structure of inactive FZD1 | | Descriptor: | Frizzled-1,Soluble cytochrome b562, anti-BRIL Fab Heavy Chain, anti-BRIL Fab Light Chain, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
2YSF
 
 | | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH
To be Published
|
|
6I4I
 
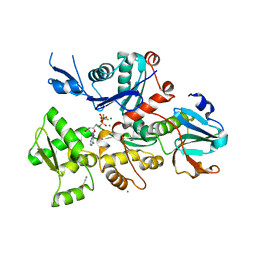 | | Crystal Structure of Plasmodium falciparum actin I (F54Y mutant) in the Mg-K-ADP-AlFn state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Actin-1, ... | | Authors: | Kumpula, E.-P, Lopez, A.J, Tajedin, L, Han, H, Kursula, I. | | Deposit date: | 2018-11-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic view into Plasmodium actin polymerization, ATP hydrolysis, and fragmentation.
Plos Biol., 17, 2019
|
|
3KFP
 
 | |
2PON
 
 | | Solution structure of the Bcl-xL/Beclin-1 complex | | Descriptor: | Apoptosis regulator Bcl-X, Beclin-1 | | Authors: | Feng, W, Huang, S, Wu, H, Zhang, M. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Bcl-xL's Target Recognition Versatility Revealed by the Structure of Bcl-xL in Complex with the BH3 Domain of Beclin-1.
J.Mol.Biol., 372, 2007
|
|
8JMI
 
 | |
2Q0R
 
 | | Structure of Pectenotoxin-2 Bound to Actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, CALCIUM ION, ... | | Authors: | Allingham, J.S, Miles, C.O, Rayment, I. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural basis for regulation of actin polymerization by pectenotoxins.
J.Mol.Biol., 371, 2007
|
|
2PQK
 
 | | X-ray crystal structure of human Mcl-1 in complex with Bim BH3 | | Descriptor: | Bim BH3 peptide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Bare, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KG0
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
6IJO
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
8J8K
 
 | | Membrane bound PRTase, C3 symmetry, acceptor bound | | Descriptor: | Decaprenyl-phosphate phosphoribosyltransferase, MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE | | Authors: | Wu, F.Y, Gao, S, Zhang, L, Rao, Z.H. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis.
Nat Microbiol, 9, 2024
|
|
1UZE
 
 | | Complex of the anti-hypertensive drug enalaprilat and the human testicular angiotensin I-converting enzyme | | Descriptor: | 1-((2S)-2-{[(1S)-1-CARBOXY-3-PHENYLPROPYL]AMINO}PROPANOYL)-L-PROLINE, ANGIOTENSIN CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Natesh, R, Schwager, S.L.U, Evans, H.R, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2004-03-11 | | Release date: | 2004-07-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Details on the Binding of Antihypertensive Drugs Captopril and Enalaprilat to Human Testicular Angiotensin I-Converting Enzyme
Biochemistry, 43, 2004
|
|
1US2
 
 | | Xylanase10C (mutant E385A) from Cellvibrio japonicus in complex with xylopentaose | | Descriptor: | ENDO-BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
