6J1S
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SS | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Lipase B, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
7VW6
 
 | | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoshikawa, T, Makino, F, Miyata, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Chem.Commun.(Camb.), 58, 2022
|
|
6VEI
 
 | | Crystal Structure of Human Cytosolic Isocitrate Dehydrogenase (IDH1) R132H Mutant in Complex with NADPH and AG-881 (Vorasidenib) Inhibitor | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, ACETATE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
4UR0
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans in complex with trichloroethene | | Descriptor: | 1,1,2-trichloroethene, BENZAMIDINE, GLYCEROL, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
6V3R
 
 | | Crystal structure of murine cycloxygenase in complex with a harmaline analog, 4,9-dihydro-3H-pyrido[3,4-b]indole | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-[(4-chlorophenyl)methyl]-6-methoxy-1-methyl-4,9-dihydro-3H-beta-carboline, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Harmaline Analogs as Substrate-Selective Cyclooxygenase-2 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
4WEO
 
 | |
6JD8
 
 | |
4V4C
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4TZT
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS ENOYL REDUCTASE (INHA) COMPLEXED WITH N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL- 5-OXOPYRROLIDINE-3-CARBOXAMIDE | | Descriptor: | (3S)-N-(3-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4U6T
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a at 1.76 Angstrom resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6JHF
 
 | | Crystal structure of apo Pullulanase from Paenibacillus barengoltzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6J1P
 
 | | Crystal structure of Candida Antarctica Lipase B mutant - SR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
4U6Q
 
 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | Descriptor: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
6J1Q
 
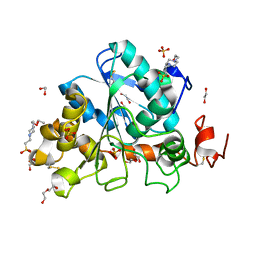 | | Crystal structure of Candida Antarctica Lipase B mutant - RS | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cen, Y.X, Zhou, J.H, Wu, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereodivergent Protein Engineering of a Lipase To Access All Possible Stereoisomers of Chiral Esters with Two Stereocenters.
J.Am.Chem.Soc., 141, 2019
|
|
7W5Z
 
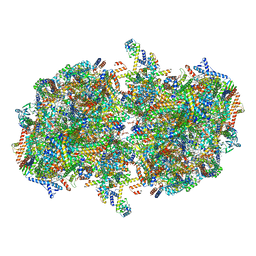 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial complex IV, composite dimer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-oxoglutarate/malate carrier protein, CARDIOLIPIN, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Letts, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
6JK6
 
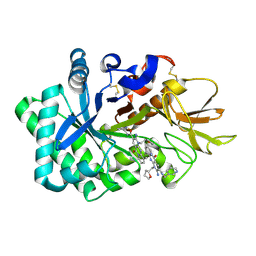 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 2-8-s2 | | Descriptor: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitotriosidase-1 | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-27 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JD1
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADH, and CPD at pH7.5 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JMW
 
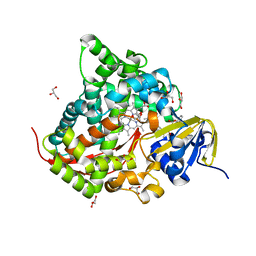 | | Structure of the Chromium Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, Chromium Protoporphyrin IX, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6J3X
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 with Maltotriose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucan 1,4-alpha-maltotetraohydrolase, ... | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2019-01-06 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Maltotetraose-forming amylase from Pseudomonas saccharophila STB07
To Be Published
|
|
6JLV
 
 | | Near-Atomic Resolution Structure of the CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7VVR
 
 | | Bovine cytochrome c oxidese in CN-bound mixed valence state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7VUW
 
 | | Bovine heart cytochrome c oxidase in the cyanide-bound fully oxidized state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
6JMH
 
 | | Structure of the Oxomolybdenum Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-10 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6IK6
 
 | | Crystal structure of Tomato beta-galactosidase (TBG) 4 with beta-1,4-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
