7WU0
 
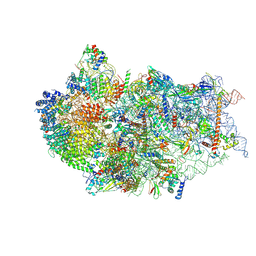 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B3 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTL
 
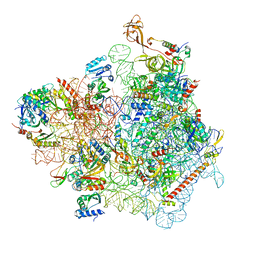 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Dis-D | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, La Venuta, G, Lau, B, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In vitro structural maturation of an early stage pre-40S particle coupled with U3 snoRNA release and central pseudoknot formation.
Nucleic Acids Res., 50, 2022
|
|
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The global structure of dormant ribosome
To Be Published
|
|
8GW7
 
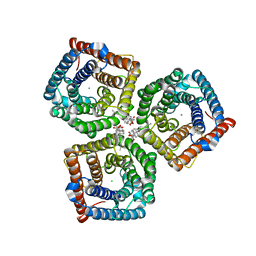 | | AtSLAC1 6D mutant in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8GW6
 
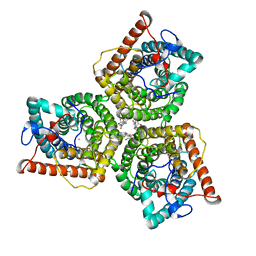 | | AtSLAC1 6D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein (Fragment) | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2022-09-16 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8T6L
 
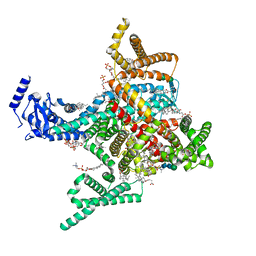 | | Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B | | Descriptor: | (1R)-1-[(5aR,7aR,9R,11aS,11bS,12R,13aR)-9,12-dihydroxy-2,11a-dimethyl-1,2,3,4,7a,8,9,10,11,11a,12,13-dodecahydro-7H-9,11b-epoxy-13a,5a-prop[1]enophenanthro[2,1-f][1,4]oxazepin-14-yl]ethyl benzoate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Tonggu, L, Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dual receptor-sites reveal the structural basis for hyperactivation of sodium channels by poison-dart toxin batrachotoxin.
Nat Commun, 15, 2024
|
|
7V87
 
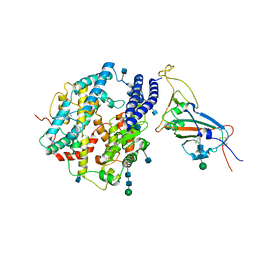 | | Local refinement of SARS-CoV-2 S-Kappa variant (B.1.617.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Local refinement of SARS-CoV-2 S-Kappa variant (B.1.617.1) RBD and Angiotensin-converting enzyme 2 (ACE2) ectodomain
To Be Published
|
|
7V85
 
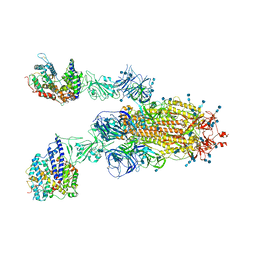 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form
To Be Published
|
|
7K18
 
 | | Cardiac Sodium channel with toxin bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-like toxin Lqh3, ... | | Authors: | Jiang, D, Catterall, W.A. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for voltage-sensor trapping of the cardiac sodium channel by a deathstalker scorpion toxin.
Nat Commun, 12, 2021
|
|
7V88
 
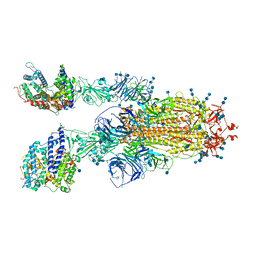 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, two ACE2-bound form
To Be Published
|
|
7EDJ
 
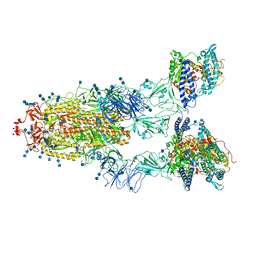 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-01 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5TGA
 
 | |
5M1J
 
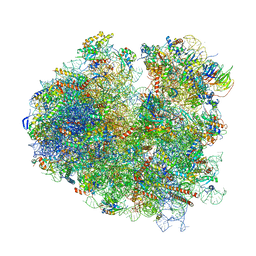 | | Nonstop ribosomal complex bound with Dom34 and Hbs1 | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Hilal, T, Yamamoto, H, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution.
Nat Commun, 7, 2016
|
|
7AH9
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
8OTS
 
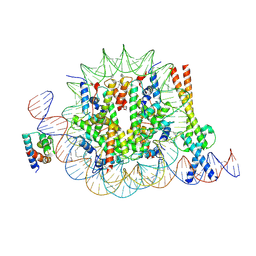 | | OCT4 and MYC-MAX co-bound to a nucleosome | | Descriptor: | DNA (127-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
6SWC
 
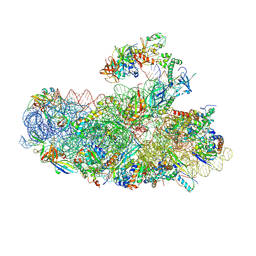 | | IC2B model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
4U5T
 
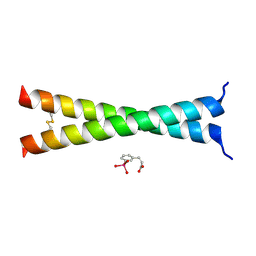 | | Crystal Structure of VBP Leucine Zipper with Bound Arylstibonic Acid | | Descriptor: | (2Z)-3-{3-[dihydroxy(oxido)-lambda~5~-stibanyl]phenyl}prop-2-enoic acid, VBP leucine zipper | | Authors: | Stagno, J.R, Ji, X. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | P6981, an arylstibonic acid, is a novel low nanomolar inhibitor of cAMP response element-binding protein binding to DNA.
Mol.Pharmacol., 82, 2012
|
|
5LZS
 
 | | Structure of the mammalian ribosomal elongation complex with aminoacyl-tRNA, eEF1A, and didemnin B | | Descriptor: | (2~{S})-~{N}-[(2~{R})-1-[[(3~{S},6~{S},8~{S},12~{S},13~{R},16~{S},17~{R},20~{S},23~{S})-13-[(2~{S})-butan-2-yl]-20-[(4-methoxyphenyl)methyl]-6,17,21-trimethyl-3-(2-methylpropyl)-12-oxidanyl-2,5,7,10,15,19,22-heptakis(oxidanylidene)-8-propan-2-yl-9,18-dioxa-1,4,14,21-tetrazabicyclo[21.3.0]hexacosan-16-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-~{N}-methyl-1-[(2~{S})-2-oxidanylpropanoyl]pyrrolidine-2-carboxamide, 18S ribosomal RNA, 28S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
7UIW
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7XHB
 
 | |
8BRM
 
 | | Giardia ribosome in POST-T state, no E-site tRNA (A6) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-23 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
5XXU
 
 | | Small subunit of Toxoplasma gondii ribosome | | Descriptor: | 18S RNA, Ribosomal protein eL41, Ribosomal protein eS1, ... | | Authors: | LI, Z, Guo, Q, Zheng, L, Ji, Y, Xie, Y, Lai, D, Lun, Z, Suo, X, Gao, N. | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-30 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii
Cell Res., 27, 2017
|
|
6WV9
 
 | |
6WVA
 
 | |
8T1D
 
 | | Open-state cryo-EM structure of full-length human TRPV4 in complex with agonist 4a-PDD | | Descriptor: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
