6PIA
 
 | |
6MHF
 
 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6G4G
 
 | | Full length ectodomain of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) including the SMB domains but with a partially disordered active site structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dohler, C, Zebisch, M, Strater, N. | | Deposit date: | 2018-03-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallization of ectonucleotide phosphodiesterase/pyrophosphatase-3 and orientation of the SMB domains in the full-length ectodomain.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6PJ7
 
 | |
6Y5O
 
 | | The crystal structure of glycogen phosphorylase in complex with 20 | | Descriptor: | 2-(4-fluorophenyl)-5,7-bis(oxidanyl)chromen-4-one, Glycogen phosphorylase, muscle form | | Authors: | Kyriakis, E, Koulas, S.M, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Synthetic flavonoid derivatives targeting the glycogen phosphorylase inhibitor site: QM/MM-PBSA motivated synthesis of substituted 5,7-dihydroxyflavones, crystallography, in vitro kinetics and ex-vivo cellular experiments reveal novel potent inhibitors.
Bioorg.Chem., 102, 2020
|
|
6VBZ
 
 | | Crystal structure of the rat MLKL pseudokinase domain | | Descriptor: | MANGANESE (II) ION, Mixed lineage kinase domain-like pseudokinase | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2019-12-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues.
Nat Commun, 11, 2020
|
|
8KFQ
 
 | |
6YA6
 
 | | Minimal construct of Cdc7-Dbf4 bound to XL413 | | Descriptor: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, BORIC ACID, CHLORIDE ION, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
6MJE
 
 | |
5DJ7
 
 | |
6PBJ
 
 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase with Gly190Pro mutation | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Jiao, W, Fan, Y, Blackmore, N.J, Parker, E.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A single amino acid substitution uncouples catalysis and allostery in an essential biosynthetic enzyme in Mycobacterium tuberculosis .
J.Biol.Chem., 295, 2020
|
|
6G66
 
 | |
7CC2
 
 | |
6PJV
 
 | | Structure of Human Sonic Hedgehog in complex with Zinc and Magnesium | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Bonn-Breach, R.B, Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of Sonic Hedgehog protein in complex with zinc(II) and magnesium(II) reveals ion-coordination plasticity relevant to peptide drug design.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6G6B
 
 | |
6G6F
 
 | |
6G6I
 
 | | MamM CTD W247A | | Descriptor: | BETA-MERCAPTOETHANOL, Magnetosome protein MamM | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-04-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to the dynamic cytoplasmic domain of the cation diffusion facilitator (CDF) protein MamM induces a 'locked-in' configuration.
Febs J., 286, 2019
|
|
7CB4
 
 | | Crystal structures of of BlAsnase | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
6PBR
 
 | | Catalytic domain of E.coli dihydrolipoamide succinyltransferase in I4 space group | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, SODIUM ION | | Authors: | Andi, B, Soares, A.S, Shi, W, Fuchs, M.R, McSweeney, S, Liu, Q. | | Deposit date: | 2019-06-14 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the dihydrolipoamide succinyltransferase catalytic domain from Escherichia coli in a novel crystal form: a tale of a common protein crystallization contaminant.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6MKA
 
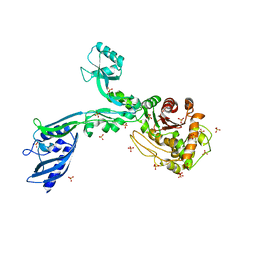 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the open conformation | | Descriptor: | SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
5DH1
 
 | |
8KH3
 
 | |
6MRV
 
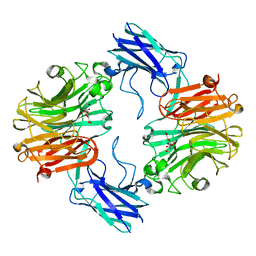 | | Sialidase26 co-crystallized with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6G6N
 
 | | Crystal structure of the computationally designed Tako8 protein in C2 | | Descriptor: | Tako8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
7C9B
 
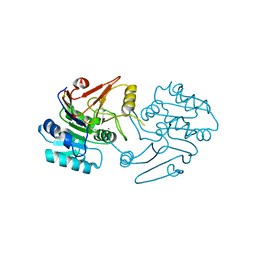 | | Crystal structure of dipeptidase-E from Xenopus laevis | | Descriptor: | Alpha-aspartyl dipeptidase, CALCIUM ION, SODIUM ION | | Authors: | Kumar, A, Singh, R, Makde, R.D. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of aspartyl dipeptidase from Xenopus laevis revealed ligand binding induced loop ordering and catalytic triad assembly.
Proteins, 90, 2022
|
|
