8PXH
 
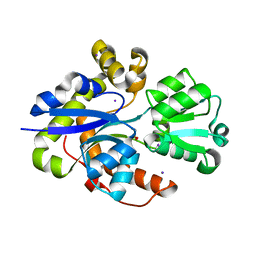 | | Structure of TauA from E. coli, solved at wavelength 2.375 A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, IODIDE ION, Taurine ABC transporter substrate-binding protein | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX7
 
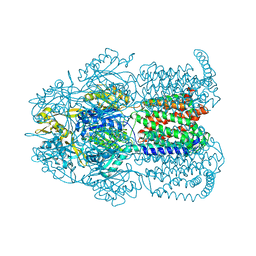 | | Structure of Bacterial Multidrug Efflux transporter AcrB, solved at wavelength 3.02 A | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
5USV
 
 | |
6N4Y
 
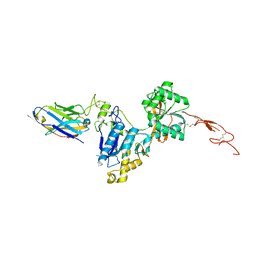 | | Metabotropic Glutamate Receptor 5 Extracellular Domain with Nb43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Mathiesen, J.M, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.262 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
7OFX
 
 | | Crystal structure of a GH31 family sulfoquinovosidase mutant D455N from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | Alpha-glucosidase yihQ, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5LGJ
 
 | |
6MVR
 
 | | Structure of a bacterial ALDH16 | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Tanner, J.J, Liu, L. | | Deposit date: | 2018-10-28 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase 16 Reveals Trans-Hierarchical Structural Similarity and a New Dimer.
J. Mol. Biol., 431, 2019
|
|
8Q58
 
 | |
7QXZ
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 5 | | Descriptor: | 2-[(3S)-1-[[2-[3,5-bis(chloranyl)phenyl]-6-[2-(4-methylpiperazin-4-ium-1-yl)pyrimidin-5-yl]oxy-pyridin-4-yl]methyl]pyrrolidin-1-ium-3-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7WF9
 
 | |
5LGW
 
 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant co-crystallised with maltodextrin | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
8Q5A
 
 | | Crystal structure of metal-dependent class II sulfofructosephosphate aldolase from Hafnia paralvei HpSqiA-Zn in complex with dihydroxyacetone phosphate (DHAP) | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Ketose-bisphosphate aldolase, ZINC ION | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
7W07
 
 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate, Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Sun, P.K, Wang, B, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
5LHY
 
 | | PB3 Domain of Human PLK4 (apo) | | Descriptor: | Serine/threonine-protein kinase PLK4 | | Authors: | Cottee, M.A, Johnson, S, Lea, S.M. | | Deposit date: | 2016-07-13 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | A key centriole assembly interaction interface between human PLK4 and STIL appears to not be conserved in flies.
Biol Open, 6, 2017
|
|
6N5U
 
 | | Crystal structure of Arabidopsis thaliana ScoI with copper bound | | Descriptor: | COPPER (I) ION, Protein SCO1 homolog 1, mitochondrial | | Authors: | Lisa, M.N, Giannini, E, Llases, M.E, Alzari, P.M, Vila, A.J. | | Deposit date: | 2018-11-22 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Arabidopsis thaliana Hcc1 is a Sco-like metallochaperone for CuAassembly in Cytochrome c Oxidase.
Febs J., 287, 2020
|
|
7QK7
 
 | | Crystal structure of the APO form of ALDH1A3 | | Descriptor: | Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
8Q57
 
 | | Crystal structure of class II SFP aldolase from Yersinia aldovae (YaSqiA-Zn-SO4) with bound sulfate ions | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
7W5E
 
 | | Oxidase ChaP D49L mutant | | Descriptor: | ChaP, FE (III) ION | | Authors: | Wang, Y, Zheng, W, Meng, Z, Jin, Y, Zhu, J, Tan, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Alteration of the Catalytic Reaction Trajectory of a Vicinal Oxygen Chelate Enzyme by Directed Evolution.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5LH9
 
 | |
6MY5
 
 | |
6N6R
 
 | |
8DF8
 
 | |
7W61
 
 | | Crystal structure of farnesol dehydrogenase from Helicoverpa armigera | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Farnesol dehydrogenase, ... | | Authors: | Kumar, R, Das, J, Mahto, J.K, Sharma, M, Kumar, P, Sharma, A.K. | | Deposit date: | 2021-12-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and molecular characterization of NADP + -farnesol dehydrogenase from cotton bollworm, Helicoverpaarmigera.
Insect Biochem.Mol.Biol., 147, 2022
|
|
7QK8
 
 | | Crystal structure of the ALDH1A3-NAD+ complex | | Descriptor: | Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
7QXY
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 3 | | Descriptor: | 3-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyrimidin-2-yl]piperazin-1-ium-1-yl]propanoate, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
