8XMN
 
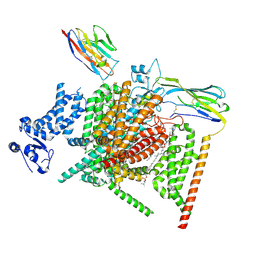 | | Voltage-gated sodium channel Nav1.7 variant M2 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XMO
 
 | | Voltage-gated sodium channel Nav1.7 variant M4 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4LQI
 
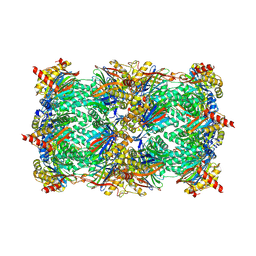 | | Yeast 20S Proteasome in complex with Vibralactone | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | List, A, Zeiler, E, Gallastegui, N, Rusch, M, Hedberg, C, Sieber, S.A, Groll, M. | | Deposit date: | 2013-07-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Omuralide and Vibralactone: Differences in the Proteasome-beta-Lactone-gamma-Lactam Binding Scaffold Alter Target Preferences.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6AAO
 
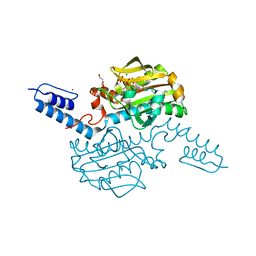 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TCO*Lys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
4O4J
 
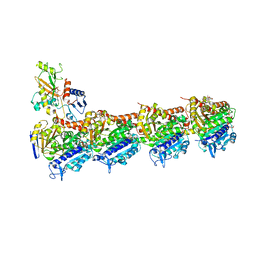 | | Tubulin-Peloruside A complex | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside A.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4O4H
 
 | | Tubulin-Laulimalide complex | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside a.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4O4L
 
 | | Tubulin-Peloruside A-Epothilone A complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, EPOTHILONE A, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside a.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
7RQW
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an open state, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
4NAB
 
 | | Structure of the (SR)Ca2+-ATPase mutant E309Q in the Ca2-E1-MgAMPPCP form | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Bublitz, M, Clausen, J.D, Arnou, B, Montigny, C, Jaxel, C, Nissen, P, Moller, J.V, Andersen, J.P, le Maire, M. | | Deposit date: | 2013-10-22 | | Release date: | 2013-12-18 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | SERCA mutant E309Q binds two Ca(2+) ions but adopts a catalytically incompetent conformation.
Embo J., 32, 2013
|
|
9EZ1
 
 | | Vitamin D receptor in complex with 1,4a,25-trihydroxyvitamin D3 | | Descriptor: | 1,4a,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
5X8S
 
 | | Crystal Structure of the mutant Human ROR gamma Ligand Binding Domain With Ursolic acid. | | Descriptor: | Nuclear receptor ROR-gamma, Ursolic acid | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
9BJK
 
 | | Inactive mu opioid receptor bound to Nb6, naloxone and NAM | | Descriptor: | Mu-type opioid receptor, Naloxone, Nalpha-[({(1M)-1-[5-(benzyloxy)pyridin-3-yl]naphthalen-2-yl}sulfanyl)acetyl]-3-methoxy-N,4-dimethyl-L-phenylalaninamide, ... | | Authors: | O'Brien, E.S, Wang, H, Kaavya Krishna, K, Zhang, C, Kobilka, B.K. | | Deposit date: | 2024-04-25 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | A mu-opioid receptor modulator that works cooperatively with naloxone.
Nature, 631, 2024
|
|
8WA5
 
 | |
9BC7
 
 | | HCN1 M305L holo | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
9BC6
 
 | | HCN1 M305L with propofol | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,6-BIS(1-METHYLETHYL)PHENOL, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Kim, E.D, Nimigean, C.M. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants.
Nature, 632, 2024
|
|
5YLU
 
 | | Crystal structure of the gastric proton pump complexed with vonoprazan | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[5-(2-fluorophenyl)-1-pyridin-3-ylsulfonyl-pyrrol-3-yl]-~{N}-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Fujiyoshi, Y. | | Deposit date: | 2017-10-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79988956 Å) | | Cite: | Crystal structures of the gastric proton pump
Nature, 556, 2018
|
|
4O4I
 
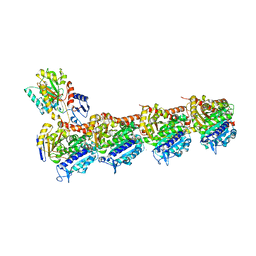 | | Tubulin-Laulimalide-Epothilone A complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, EPOTHILONE A, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside a.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3HD3
 
 | | High resolution crystal structure of cruzain bound to the vinyl sulfone inhibitor SMDC-256047 | | Descriptor: | (1R,2R)-2-[(4-chlorophenyl)carbonyl]-N-{(1S)-1-[2-(phenylsulfonyl)ethyl]pentyl}cyclohexanecarboxamide, 1,2-ETHANEDIOL, Cruzipain, ... | | Authors: | Kerr, I.D, Debnath, M, Brinen, L.S. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-06 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel non-peptidic vinylsulfones targeting the S2 and S3 subsites of parasite cysteine proteases.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IRX
 
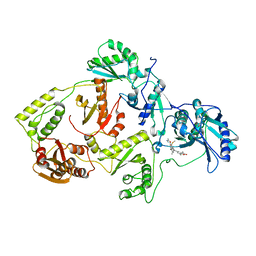 | | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the Non-nucleoside RT Inhibitor (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate. | | Descriptor: | (E)-S-Methyl 5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methy lbenzothioate, Reverse transcriptase, Reverse transcriptase/ribonuclease H | | Authors: | Ho, W.C, Arnold, E. | | Deposit date: | 2009-08-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 reverse transcriptase (RT) in complex with the
alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor (E)-S-Methyl
5-(1-(3,7-Dimethyl-2-oxo-2,3-dihydrobenzo[d]oxazol-5-yl)-5-(5-methyl-1,3,4-oxadiazol-2-yl)pent-1-enyl)-2-methoxy-3-methylbenzothioate.
J.Med.Chem., 52, 2009
|
|
3OGL
 
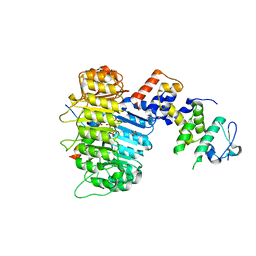 | | Structure of COI1-ASK1 in complex with JA-isoleucine and the JAZ1 degron | | Descriptor: | Coronatine-insensitive protein 1, JAZ1 incomplete degron peptide, N-({(1R,2S)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetyl)-L-isoleucine, ... | | Authors: | Sheard, L.B, Tan, X, Mao, H, Withers, J, Ben-Nissan, G, Hinds, T.R, Hsu, F, Sharon, M, Browse, J, He, S.Y, Rizo, J, Howe, G.A, Zheng, N. | | Deposit date: | 2010-08-17 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor.
Nature, 468, 2010
|
|
6O9G
 
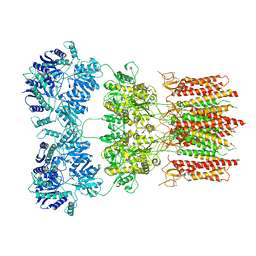 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
6O9U
 
 | | KirBac3.1 at a resolution of 2 Angstroms | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | Authors: | Gulbis, J.M, Clarke, O.B. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6OAM
 
 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2, Ubiquitin | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2019-03-17 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
3K52
 
 | |
