7NBW
 
 | |
1C9A
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-11-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
4OAS
 
 | | co-crystal structure of MDM2 (17-111) in complex with compound 25 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2014-01-06 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of AMG 232, a Potent, Selective, and Orally Bioavailable MDM2-p53 Inhibitor in Clinical Development.
J.Med.Chem., 57, 2014
|
|
4OVG
 
 | | E. coli sliding clamp in complex with (R)-9-(2-amino-2-oxoethyl)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-9-(2-amino-2-oxoethyl)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4ODE
 
 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | (2-{[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]methyl}-1,3-thiazol-5-yl)acetic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4D7D
 
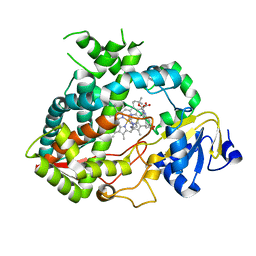 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-({3-oxo-3-[(pyridin-3-ylmethyl)amino]propyl}sulfanyl)propan-2-yl]carbamate | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-22 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
4D78
 
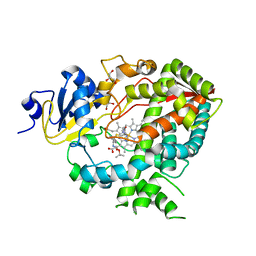 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-21 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
6E3E
 
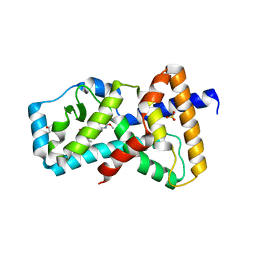 | | Structure of RORgt in complex with a novel inverse agonist. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(5R)-5-[(7-fluoro-1,1-dimethyl-2,3-dihydro-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5H)-yl]-5-oxopentanoic acid, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
6E3G
 
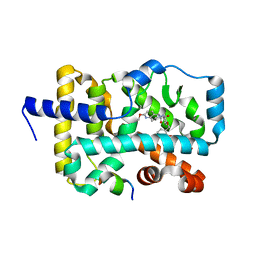 | | Structure of RORgt in complex with a novel agonist. | | Descriptor: | (5R)-6-acetyl-2-methoxy-N-{4-[(2-methoxyphenyl)methoxy]phenyl}-5,6,7,8-tetrahydro-1,6-naphthyridine-5-carboxamide, 1,2-ETHANEDIOL, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
4L3P
 
 | | Crystal Structure of 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine bound to TAK1-TAB1 | | Descriptor: | 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Steinbacher, S, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4L52
 
 | | Crystal Structure of 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethan-1-one bound to TAK1-TAB1 | | Descriptor: | 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethanone, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4DZB
 
 | |
2JC0
 
 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB655264 | | Descriptor: | (2S,4S,5R)-2-ISOBUTYL-5-(2-THIENYL)-1-[4-(TRIFLUOROMETHYL)BENZOYL]PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
2JC1
 
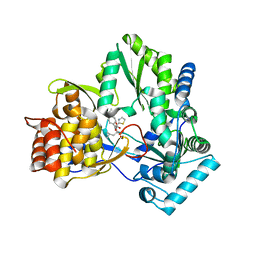 | | CRYSTAL STRUCTURE OF HEPATITIS C VIRUS POLYMERASE IN COMPLEX WITH INHIBITOR SB698223 | | Descriptor: | (2S,4S,5R)-1-(4-TERT-BUTYLBENZOYL)-2-ISOBUTYL-5-(1,3-THIAZOL-2-YL)PYRROLIDINE-2,4-DICARBOXYLIC ACID, RNA-DEPENDENT RNA-POLYMERASE | | Authors: | Wonacott, A, Skarzynski, T, Singh, O.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of Novel Acyl Pyrrolidine Inhibitors of Hepatitis C Virus RNA-Dependent RNA Polymerase Leading to a Development Candidate.
J.Med.Chem., 50, 2007
|
|
4D6Z
 
 | |
4D75
 
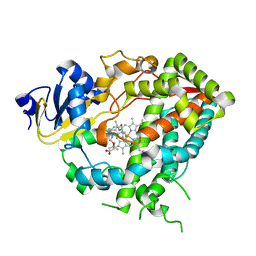 | | Cytochrome P450 3A4 bound to an inhibitor | | Descriptor: | CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl {6-oxo-6-[(pyridin-3-ylmethyl)amino]hexyl}carbamate | | Authors: | Sevrioukova, I, Poulos, T. | | Deposit date: | 2014-11-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Inhibitor Design for Evaluation of a Cyp3A4 Pharmacophore Model.
J.Med.Chem., 59, 2016
|
|
8TXT
 
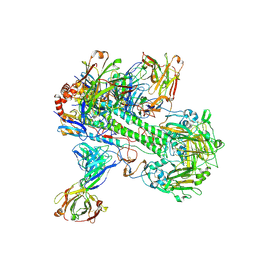 | | Crystal structure of 05.GC.w13.02 Fab in complex with H5 HA from A/Viet Nam/1203/2004(H5N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GC_W13B_B, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
8TY7
 
 | | Crystal structure of 05.GC.w2.3C10 Fab in complex with H1 HA from A/California/04/2009(H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GC_w2_3C10, heavy chain, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
8TXP
 
 | | Crystal structure of 05.GC.w13.01 Fab in complex with H1 HA from A/California/04/2009(H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GC_w13_A, Fab heavy chain, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
8TXM
 
 | | Crystal structure of 05.GC.w13.02 Fab in complex with H1 HA from A/California/04/2009(H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GC_w13_B, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4JEF
 
 | |
7OKQ
 
 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | Descriptor: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | Authors: | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
3QJZ
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
