8WH5
 
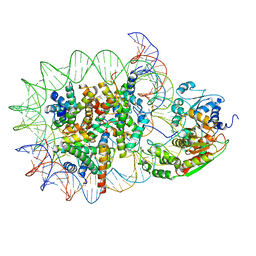 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGU
 
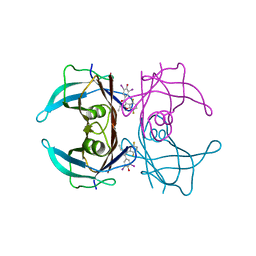 | | Crystal structure of V30M-TTR in complex with compound 20 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-[2-ethyl-4,7-bis(fluoranyl)-1-benzofuran-3-yl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGT
 
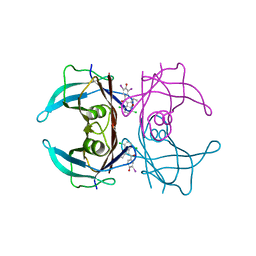 | | Crystal structure of V30M-TTR in complex with compound 7 | | Descriptor: | Transthyretin, [4,7-bis(chloranyl)-2-ethyl-1-benzofuran-3-yl]-[3,5-bis(iodanyl)-4-oxidanyl-phenyl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGS
 
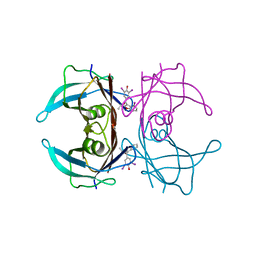 | | Crystal structure of V30M-TTR in complex with compound 4 | | Descriptor: | Transthyretin, [3,5-bis(iodanyl)-4-oxidanyl-phenyl]-(2-ethyl-4-iodanyl-1-benzofuran-3-yl)methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
8WGQ
 
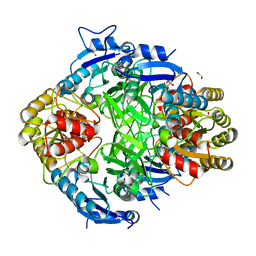 | | The Crystal Structure of L-asparaginase from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-asparaginase | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2023-09-22 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of L-asparaginase from Biortus.
To Be Published
|
|
8WGP
 
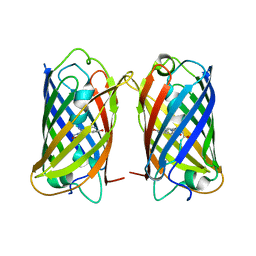 | | Crystal structure of DsRed-Monomer | | Descriptor: | Red fluorescent protein | | Authors: | Nam, K.H. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Flexibility of the Monomeric Red Fluorescent Protein DsRed.
Crystals, 14, 2024
|
|
8WGL
 
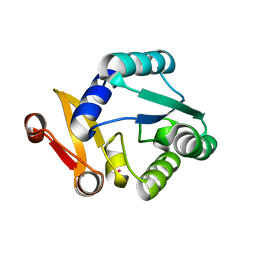 | | Crystal structure of Rhodothermus marinus substrate-binding protein (Hg soaking) | | Descriptor: | ABC-type uncharacterized transport system periplasmic component-like protein, MERCURY (II) ION | | Authors: | Nam, K.H. | | Deposit date: | 2023-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and bioinformatics analysis of single-domain substrate-binding protein from Rhodothermus marinus.
Biochem Biophys Rep, 37, 2024
|
|
8WGK
 
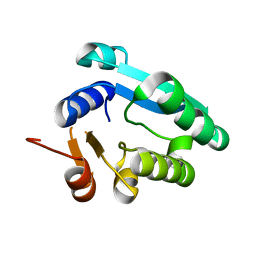 | |
8WGH
 
 | | Cryo-EM structure of the red-shifted Fittonia albivenis PSI-LHCI | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Huang, G.Q, Li, X.X, Sui, S.F, Qin, X.C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structure of the red-shifted Fittonia albivenis PSI-LHCI
To Be Published
|
|
8WGF
 
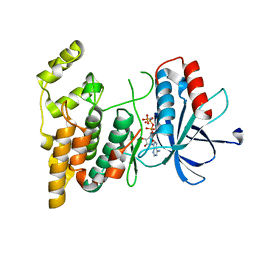 | | The Crystal Structure of JNK3 from Biortus. | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 10, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2023-09-21 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of JNK3 from Biortus.
To Be Published
|
|
8WG8
 
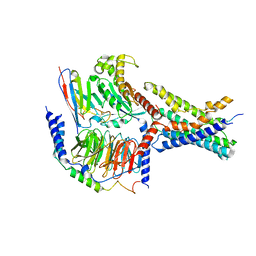 | | Cryo-EM structures of peptide free and Gs-coupled GCGR | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8WG7
 
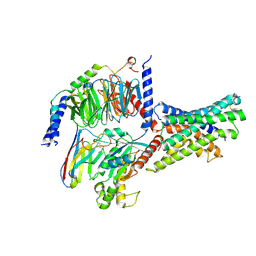 | | Cryo-EM structures of peptide free and Gs-coupled GLP-1R | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8WG5
 
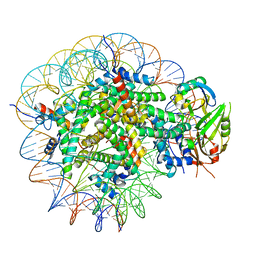 | | Cryo-EM structure of USP16 bound to H2AK119Ub nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, He, Z.Z, Deng, Z.H, Liu, L. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and mechanistic basis for nucleosomal H2AK119 deubiquitination by single-subunit deubiquitinase USP16.
Nat.Struct.Mol.Biol., 2024
|
|
8WG2
 
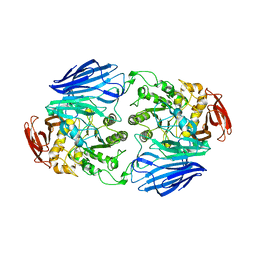 | |
8WG1
 
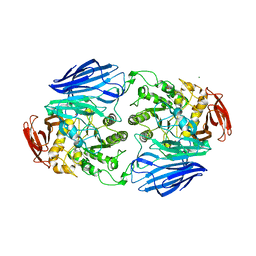 | | Crystal structure of GH97 glucodextranase mutant E509Q from Flavobacterium johnsoniae in complex with panose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural insights into alpha-(1->6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 2024
|
|
8WG0
 
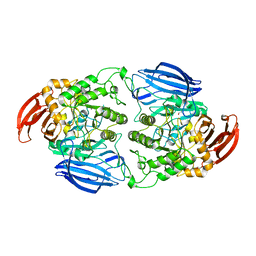 | | Crystal structure of GH97 glucodextranase from Flavobacterium johnsoniae in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into alpha-(1->6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 2024
|
|
8WFY
 
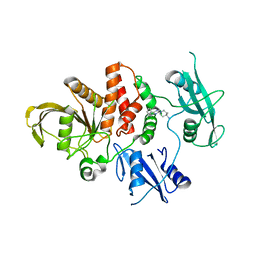 | | The Crystal Structure of SHP2 from Biortus. | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of SHP2 from Biortus.
To Be Published
|
|
8WFW
 
 | |
8WFV
 
 | |
8WFU
 
 | |
8WFT
 
 | |
8WFR
 
 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
8WFQ
 
 | | The Crystal Structure of RALDH1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde dehydrogenase 1A1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Shen, Z. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of RALDH1 from Biortus.
To Be Published
|
|
